Detection and identification of bacterial endosymbionts in arbuscular mycorrhizal fungi belonging to the family Gigasporaceae
- PMID: 11010905
- PMCID: PMC92331
- DOI: 10.1128/AEM.66.10.4503-4509.2000
Detection and identification of bacterial endosymbionts in arbuscular mycorrhizal fungi belonging to the family Gigasporaceae
Abstract
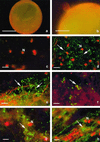
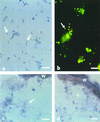
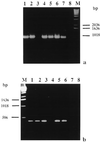
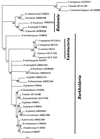
Intracellular bacteria have been found previously in one isolate of the arbuscular mycorrhizal (AM) fungus Gigaspora margarita BEG 34. In this study, we extended our investigation to 11 fungal isolates obtained from different geographic areas and belonging to six different species of the family Gigasporaceae. With the exception of Gigaspora rosea, isolates of all of the AM species harbored bacteria, and their DNA could be PCR amplified with universal bacterial primers. Primers specific for the endosymbiotic bacteria of BEG 34 could also amplify spore DNA from four species. These specific primers were successfully used as probes for in situ hybridization of endobacteria in G. margarita spores. Neighbor-joining analysis of the 16S ribosomal DNA sequences obtained from isolates of Scutellospora persica, Scutellospora castanea, and G. margarita revealed a single, strongly supported branch nested in the genus Burkholderia.
Figures
References
-
- Amann R I, Springer N, Ludwig W, Goertz H D, Schleifer K H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991;351:161–164. - PubMed
-
- Andrade G, Mihara K L, Linderman R G, Bethlenfalvay G J. Bacteria from rhizosphere and hyphosphere soils of different arbuscular-mycorrhizal fungi. Plant Soil. 1997;192:71–79.
-
- Bago B, Bentivenga S P, Brenac V, Dodd J C, Piché Y, Simon L. Molecular analysis of Gigaspora (Glomales, Gigasporaceae) New Phytol. 1998;139:581–588.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

