A PCR assay To discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA
- PMID: 11010920
- PMCID: PMC92346
- DOI: 10.1128/AEM.66.10.4571-4574.2000
A PCR assay To discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA
Abstract
Our purpose was to develop a rapid, inexpensive method of diagnosing the source of fecal pollution in water. In previous research, we identified Bacteroides-Prevotella ribosomal DNA (rDNA) PCR markers based on analysis. These markers length heterogeneity PCR and terminal restriction fragment length polymorphism distinguish cow from human feces. Here, we recovered 16S rDNA clones from natural waters that were close phylogenetic relatives of the markers. From the sequence data, we designed specific PCR primers that discriminate human and ruminant sources of fecal contamination.
Figures
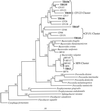
References
-
- Allsop K, Stickler J D. An assessment of Bacteroides fragilis group organisms as indicators of human faecal pollution. J Appl Bacteriol. 1985;58:95–99. - PubMed
-
- Dehority B A. Foregut fermentation. In: Mackie R I, White B A, editors. Gastrointestinal microbiology. Vol. 1. New York, N.Y: Chapman & Hall, International Thomson Publishing; 1997. p. 658.
-
- Felsenstein J. PHYLIP—phylogeny inference package (v3.5) Cladistics. 1989;5:164–166.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

