H2O2 sensing through oxidation of the Yap1 transcription factor
- PMID: 11013218
- PMCID: PMC302088
- DOI: 10.1093/emboj/19.19.5157
H2O2 sensing through oxidation of the Yap1 transcription factor
Abstract
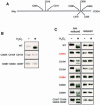
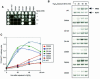
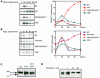
The yeast transcription factor Yap1 activates expression of antioxidant genes in response to oxidative stress. Yap1 regulation involves nuclear accumulation, but the mechanism sensing the oxidative stress signal remains unknown. We provide biochemical and genetic evidence that upon H2O2 treatment, Yap1 is activated by oxidation and deactivated by enzymatic reduction with Yap1-controlled thioredoxins, thus providing a mechanism for autoregulation. Two cysteines essential for Yap1 oxidation are also essential for its activation by H2O2. The data are consistent with a model in which oxidation of Yap1 leads to disulfide bond formation with the resulting change of conformation masking recognition of the nuclear export signal by Crm1/Xpo1, thereby promoting nuclear accumulation of the protein. In sharp contrast to H2O2, diamide does not lead to the same Yap1 oxidized form and still activates mutants lacking cysteines essential for H2O2 activation, providing a molecular basis for differential activation of Yap1 by these oxidants. This is the first example of an H2O2-sensing mechanism in a eukaryote that exploits the oxidation of cysteines in order to respond rapidly to stress conditions.
Figures

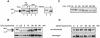


References
-
- Åslund F. and Beckwith,J. (1999) Bridge over troubled waters: sensing stress by disulfide bond formation. Cell, 96, 751–753. - PubMed
-
- Chae H.Z., Chung,S.J. and Rhee,S.G. (1994) Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem., 269, 27670–27678. - PubMed
-
- Gaudu P., Moon,N. and Weiss,B. (1997) Regulation of the soxRS oxidative stress regulon. Reversible oxidation of the Fe-S centers of SoxR in vivo. J. Biol. Chem., 272, 5082–5086. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

