Transcriptional silencing and promoter methylation triggered by double-stranded RNA
- PMID: 11013221
- PMCID: PMC302106
- DOI: 10.1093/emboj/19.19.5194
Transcriptional silencing and promoter methylation triggered by double-stranded RNA
Abstract
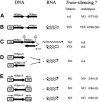
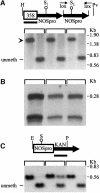
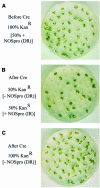
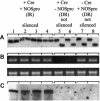
Double-stranded RNA induces a post-transcriptional gene silencing process, termed RNAi, in diverse organisms. It is shown here that transcriptional gene silencing accompanied by de novo methylation of a target promoter in plants can be triggered by a double-stranded RNA containing promoter sequences. Similar to the double-stranded RNA involved in RNAi, this promoter double-stranded RNA, which is synthesized in the nucleus, is partially cleaved into small RNAs approximately 23 nucleotides in length. Both transcriptional and post-transcriptional gene silencing can thus be initiated by double-stranded RNAs that enter the same degradation pathway. The results also implicate double-stranded RNA in directing DNA methylation. Different constructs designed to produce double-stranded promoter RNA in various ways were evaluated for their ability to induce gene silencing in tobacco and Arabidopsis. RNA hairpins transcribed from inverted DNA repeats were the most effective trans-acting silencing signals. This strategy could be useful for transcriptionally downregulating genes in a variety of plants.
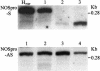
Figures



References
-
- Bass B.L. (2000) Double-stranded RNA as a template for gene silencing. Cell, 101, 235–238. - PubMed
-
- Bosher J.M. and Labouesse,M. (2000) RNA interference: genetic wand and genetic watchdog. Nature Cell Biol., 2, E31–E36. - PubMed
-
- Clough S.J. and Bent,A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J., 16, 735–743. - PubMed
-
- Cogoni C. and Macino,G. (1999a) Homology-dependent gene silencing in plants and fungi: a number of variations on the same theme. Curr. Opin. Microbiol., 2, 657–662. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

