Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm
- PMID: 11013223
- PMCID: PMC302097
- DOI: 10.1093/emboj/19.19.5212
Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm
Abstract
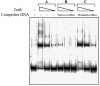
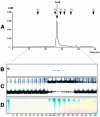
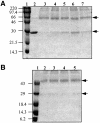
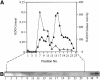
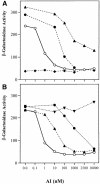
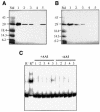
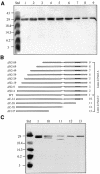
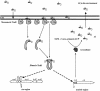
Promoter binding by TraR and LuxR, the activators of two bacterial quorum-sensing systems, requires their cognate acyl-homoserine lactone (acyl-HSL) signals, but the role the signal plays in activating these transcription factors is not known. Soluble active TraR, when purified from cells grown with the acyl-HSL, contained bound signal and was solely in dimer form. However, genetic and cross-linking studies showed that TraR is almost exclusively in monomer form in cells grown without signal. Adding signal resulted in dimerization of the protein in a concentration-dependent manner. In the absence of signal, monomer TraR localized to the inner membrane while growth with the acyl-HSL resulted in the appearance of dimer TraR in the cytoplasmic compartment. Affinity chromatography indicated that the N-terminus of TraR from cells grown without signal is hidden. Analysis of heterodimers formed between TraR and its deletion mutants localized the dimerization domain to a region between residues 49 and 156. We conclude that binding signal drives dimerization of TraR and its release from membranes into the cytoplasm.
Figures

References
-
- Adar Y.Y. and Ulitzur,S. (1993) GroESL proteins facilitate binding of externally added inducer by LuxR protein-containing E.coli cells. J. Biolumin. Chemilumin., 8, 261–266. - PubMed
-
- Cangelosi G.A., Best,E.A., Martinetti,C. and Nester,E.W. (1991) Genetic analysis of Agrobacterium tumefaciens. Methods Enzymol., 145, 177–181. - PubMed
-
- Cha C., Gao,P., Chen,Y.-C., Shaw,P.D. and Farrand,S.K. (1998) Production of acyl-homoserine lactone quorum-sensing signals by Gram-negative plant-associated bacteria. Mol. Plant Microbe Interact., 11, 1119–1129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

