Helicobacter pylori: clonal population structure and restricted transmission within families revealed by molecular typing
- PMID: 11015377
- PMCID: PMC87450
- DOI: 10.1128/JCM.38.10.3646-3651.2000
Helicobacter pylori: clonal population structure and restricted transmission within families revealed by molecular typing
Abstract
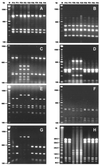
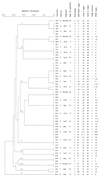
Helicobacter pylori infects up to 50% of the human population worldwide. The infection occurs predominantly in childhood and persists for decades or a lifetime. H. pylori is believed to be transmitted from person to person. However, tremendous genetic diversity has been reported for these bacteria. In order to gain insight into the epidemiological basis of this phenomenon, we performed molecular typing of H. pylori isolates from different families. Fifty-nine H. pylori isolates from 27 members of nine families were characterized by using restriction fragment length polymorphism analysis of five PCR-amplified genes, by pulsed-field gel electrophoresis (PFGE) of chromosomal DNA, and by vacA and cagA genotyping. The 16S rRNA gene exhibited little allelic variation, as expected for a unique bacterial species. In contrast, the vacA, flaA, ureAB, and lspA-glmM genes were highly polymorphic, with a mean genetic diversity of 0.83, which exceeds the levels recorded for all other bacterial species. In conjunction with PFGE, 59 H. pylori isolates could be differentiated into 21 clonal types. Each individual harbored only one clone, occasionally with a clonal variant. Identical strains were always found either between siblings or between a mother and her children. Statistical analysis revealed clonality of population structure in all isolates. The results of this study suggest the possible coexistence of a large array of clonal lineages that are evolving in each individual in isolation from one another. Transmission appears to occur primarily from mother to child and perhaps between siblings.
Figures
Similar articles
-
Genetic characterisation of Helicobacter pylori isolates from an Argentinean adult population based on cag pathogenicity island right-end motifs, lspA-glmM polymorphism and iceA and vacA genotypes.Clin Microbiol Infect. 2004 Sep;10(9):811-9. doi: 10.1111/j.1198-743X.2004.00940.x. Clin Microbiol Infect. 2004. PMID: 15355412
-
Restriction fragment length polymorphism of virulence genes cagA, vacA and ureAB of Helicobacter pylori strains isolated from Iranian patients with gastric ulcer and nonulcer disease.Saudi Med J. 2007 Apr;28(4):529-34. Saudi Med J. 2007. PMID: 17457472
-
Performance criteria of DNA fingerprinting methods for typing of Helicobacter pylori isolates: experimental results and meta-analysis.J Clin Microbiol. 1999 Dec;37(12):4071-80. doi: 10.1128/JCM.37.12.4071-4080.1999. J Clin Microbiol. 1999. PMID: 10565934 Free PMC article.
-
Molecular methods for typing of Helicobacter pylori and their applications.FEMS Immunol Med Microbiol. 1999 Jun;24(2):193-9. doi: 10.1111/j.1574-695X.1999.tb01282.x. FEMS Immunol Med Microbiol. 1999. PMID: 10378420 Review.
-
[Research progress on genotyping of Helicobacter pylori].Zhejiang Da Xue Xue Bao Yi Xue Ban. 2018 Jan 25;47(1):97-103. doi: 10.3785/j.issn.1008-9292.2018.02.14. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2018. PMID: 30146818 Free PMC article. Review. Chinese.
Cited by
-
Polymorphisms of the acid sensing histidine kinase gene arsS in Helicobacter pylori populations from anatomically distinct gastric sites.Microb Pathog. 2012 Nov-Dec;53(5-6):227-33. doi: 10.1016/j.micpath.2012.08.002. Epub 2012 Aug 30. Microb Pathog. 2012. PMID: 22940419 Free PMC article.
-
Five-year follow-up study of mother-to-child transmission of Helicobacter pylori infection detected by a random amplified polymorphic DNA fingerprinting method.J Clin Microbiol. 2005 May;43(5):2246-50. doi: 10.1128/JCM.43.5.2246-2250.2005. J Clin Microbiol. 2005. PMID: 15872250 Free PMC article.
-
Genomic methylation: a tool for typing Helicobacter pylori isolates.Appl Environ Microbiol. 2007 Jul;73(13):4243-9. doi: 10.1128/AEM.00199-07. Epub 2007 May 4. Appl Environ Microbiol. 2007. PMID: 17483255 Free PMC article.
-
Helicobacter pylori and gastroduodenal pathology: new threats of the old friend.Ann Clin Microbiol Antimicrob. 2005 Jan 5;4:1. doi: 10.1186/1476-0711-4-1. Ann Clin Microbiol Antimicrob. 2005. PMID: 15634357 Free PMC article. Review.
-
Using macro-arrays to study routes of infection of Helicobacter pylori in three families.PLoS One. 2008 May 21;3(5):e2259. doi: 10.1371/journal.pone.0002259. PLoS One. 2008. PMID: 18493595 Free PMC article.
References
-
- Achtman M, Azuma T, Berg D E, Ito Y, Morelli G, Pan Z-J, Suerbaum S, Thompson S A, van der Ende A, van Doorn L J. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–470. - PubMed
-
- Alm R A, Ling L-S L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. - PubMed
-
- Bassily S, Frenck R W, Mohareb E W, Wierzba T, Savarino S, Hall E, Kotkat A, Naficy A, Hyams K C, Clemens J. Seroprevalence of Helicobacter pylori among Egyptian newborns and their mothers: a preliminary report. Am J Trop Med Hyg. 1999;61:37–40. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

