Telomere fusions caused by mutating the terminal region of telomeric DNA
- PMID: 11016977
- PMCID: PMC17213
- DOI: 10.1073/pnas.210388397
Telomere fusions caused by mutating the terminal region of telomeric DNA
Abstract
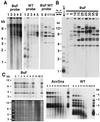
Mutations in the template region of a telomerase RNA gene can lead to the corresponding sequence alterations appearing in newly synthesized telomeric repeats. We analyzed a set of mutations in the template region of the telomerase RNA gene (TER1) of the budding yeast Kluyveromyces lactis that were predicted to lead to synthesis of mutant telomeric repeats with disrupted binding of the telomeric protein Rap1p. We showed previously that mutating the left side of the 12-bp consensus Rap1p binding site led to immediate and severe telomere elongation. Here, we show that, in contrast, mutating either the right side of the site or both sides together leads initially to telomere shortening. On additional passaging, certain mutants of both categories exhibit telomere-telomere fusions. Often, six new Bal-31-resistant, telomere repeat-containing bands appeared, and we infer that each of the six K. lactis chromosomes became circularized. These fusions were not stable, appearing occasionally to resolve and then reform. We demonstrate directly that a linear minichromosome introduced into one of the fusion mutant strains circularized by means of end-to-end fusions of the mutant repeat tracts. In contrast to the chromosomal circularization reported previously in Schizosaccharomyces pombe mutants defective in telomere maintenance, the K. lactis telomere fusions retained their telomeric DNA repeat sequences.
Figures
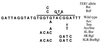


Similar articles
-
Genetic dissection of the Kluyveromyces lactis telomere and evidence for telomere capping defects in TER1 mutants with long telomeres.Eukaryot Cell. 2004 Apr;3(2):369-84. doi: 10.1128/EC.3.2.369-384.2004. Eukaryot Cell. 2004. PMID: 15075267 Free PMC article.
-
Rap1 protein regulates telomere turnover in yeast.Proc Natl Acad Sci U S A. 1998 Oct 13;95(21):12486-91. doi: 10.1073/pnas.95.21.12486. Proc Natl Acad Sci U S A. 1998. PMID: 9770512 Free PMC article.
-
Dynamics of telomeric DNA turnover in yeast.Genetics. 2002 Jan;160(1):63-73. doi: 10.1093/genetics/160.1.63. Genetics. 2002. PMID: 11805045 Free PMC article.
-
The telomere and telomerase: nucleic acid-protein complexes acting in a telomere homeostasis system. A review.Biochemistry (Mosc). 1997 Nov;62(11):1196-201. Biochemistry (Mosc). 1997. PMID: 9467842 Review.
-
The telomere and telomerase: how do they interact?Ciba Found Symp. 1997;211:2-13; discussion 15-9. doi: 10.1002/9780470515433.ch2. Ciba Found Symp. 1997. PMID: 9524748 Review.
Cited by
-
Recombinational telomere elongation promoted by DNA circles.Mol Cell Biol. 2002 Jul;22(13):4512-21. doi: 10.1128/MCB.22.13.4512-4521.2002. Mol Cell Biol. 2002. PMID: 12052861 Free PMC article.
-
Regulation of telomere structure and functions by subunits of the INO80 chromatin remodeling complex.Mol Cell Biol. 2007 Aug;27(16):5639-49. doi: 10.1128/MCB.00418-07. Epub 2007 Jun 11. Mol Cell Biol. 2007. PMID: 17562861 Free PMC article.
-
Yeast telomerase is specialized for C/A-rich RNA templates.Nucleic Acids Res. 2003 Mar 15;31(6):1646-55. doi: 10.1093/nar/gkg261. Nucleic Acids Res. 2003. PMID: 12626706 Free PMC article.
-
Shortened Infant Telomere Length Is Associated with Attention Deficit/Hyperactivity Disorder Symptoms in Children at Age Two Years: A Birth Cohort Study.Int J Mol Sci. 2022 Apr 21;23(9):4601. doi: 10.3390/ijms23094601. Int J Mol Sci. 2022. PMID: 35562991 Free PMC article.
-
Restriction of Ku translocation protects telomere ends.Nat Commun. 2025 Jul 24;16(1):6824. doi: 10.1038/s41467-025-61864-1. Nat Commun. 2025. PMID: 40707444 Free PMC article.
References
-
- Bryan T M, Cech T R. Curr Opin Cell Biol. 1999;11:318–324. - PubMed
-
- Pardue M L, DeBaryshe P G. Chromosoma. 1999;108:73–82. - PubMed
-
- Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L, Coviello G M, Wright W E, Weinrich S L, Shay J W. Science. 1994;266:2011–2015. - PubMed
-
- Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C, Morin M, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Science. 1998;279:349–352. - PubMed
-
- Vaziri H, Benchimol S. Curr Biol. 1998;8:279–282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

