A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo
- PMID: 11018020
- PMCID: PMC316978
- DOI: 10.1101/gad.831800
A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo
Abstract
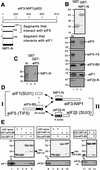
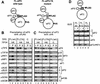
Translation initiation factor 2 (eIF2) bound to GTP transfers the initiator methionyl tRNA to the 40S ribosomal subunit. The eIF5 stimulates GTP hydrolysis by the eIF2/GTP/Met-tRNA(i)(Met) ternary complex on base-pairing between Met-tRNA(i)(Met) and the start codon. The eIF2, eIF5, and eIF1 all have been implicated in stringent selection of AUG as the start codon. The eIF3 binds to the 40S ribosome and promotes recruitment of the ternary complex; however, physical contact between eIF3 and eIF2 has not been observed. We show that yeast eIF5 can bridge interaction in vitro between eIF3 and eIF2 by binding simultaneously to the amino terminus of eIF3 subunit NIP1 and the amino-terminal half of eIF2beta, dependent on a conserved bipartite motif in the carboxyl terminus of eIF5. Additionally, the amino terminus of NIP1 can bind concurrently to eIF5 and eIF1. These findings suggest the occurrence of an eIF3/eIF1/eIF5/eIF2 multifactor complex, which was observed in cell extracts free of 40S ribosomes and found to contain stoichiometric amounts of tRNA(i)(Met). The multifactor complex was disrupted by the tif5-7A mutation in the bipartite motif of eIF5. Importantly, the tif5-7A mutant is temperature sensitive and displayed a substantial reduction in translation initiation at the restrictive temperature. We propose that the multifactor complex is an important intermediate in translation initiation in vivo.
Figures



References
-
- Asano K, Kinzy TZ, Merrick WC, Hershey JWB. Conservation and diversity of eukaryotic translation initiation factor eIF3. J Biol Chem. 1997;272:1101–1109. - PubMed
-
- Asano K, Phan L, Anderson J, Hinnebusch AG. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:18573–18585. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
