Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing
- PMID: 11018056
- PMCID: PMC2189801
- DOI: 10.1083/jcb.151.1.95
Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing
Abstract
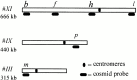
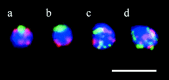
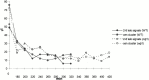
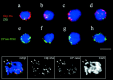
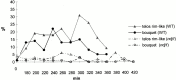
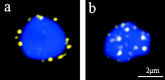
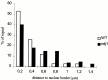
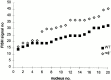
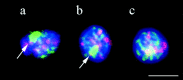
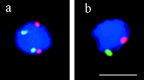
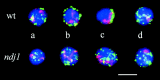
We have investigated the requirements for NDJ1 in meiotic telomere redistribution and clustering in synchronized cultures of Saccharomyces cerevisiae. On induction of wild-type meiosis, telomeres disperse from premeiotic aggregates over the nuclear periphery, and then cluster near the spindle pole body (bouquet arrangement) before dispersing again. In ndj1Delta meiocytes, telomeres are scattered throughout the nucleus and fail to form perinuclear meiosis-specific distribution patterns, suggesting that Ndj1p may function to tether meiotic telomeres to the nuclear periphery. Since ndj1Delta meiocytes fail to cluster their telomeres at any prophase stage, Ndj1p is the first protein shown to be required for bouquet formation in a synaptic organism. Analysis of homologue pairing by two-color fluorescence in situ hybridization with cosmid probes to regions on III, IX, and XI revealed that disruption of bouquet formation is associated with a significant delay (>2 h) of homologue pairing. An increased and persistent fraction of ndj1Delta meiocytes with Zip1p polycomplexes suggests that chromosome polarization is important for synapsis progression. Thus, our observations support the hypothesis that meiotic telomere clustering contributes to efficient homologue alignment and synaptic pairing. Under naturally occurring conditions, bouquet formation may allow for rapid sporulation and confer a selective advantage.
Figures

References
-
- Alani E., Padmore R., Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. - PubMed
-
- Bélar K. Chromosomenreduktion. In: Baur E., Hartmann M., editors. Handbuch der Vererbungswissenschaft, die Cytologischen Grundlagen der Vererbung. Geb. Borntraeger; Berlin: 1928. pp. 168–201.
-
- Blackburn E.H. Structure and function of telomeres. Nature. 1991;350:569–573. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

