Adeno-associated virus RNAs appear in a temporal order and their splicing is stimulated during coinfection with adenovirus
- PMID: 11024114
- PMCID: PMC102024
- DOI: 10.1128/jvi.74.21.9878-9888.2000
Adeno-associated virus RNAs appear in a temporal order and their splicing is stimulated during coinfection with adenovirus
Abstract
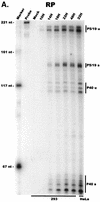
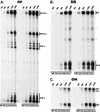
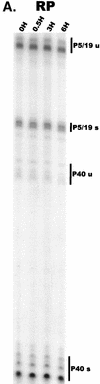
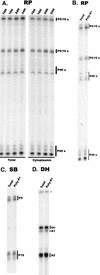
We have used a quantitative RNase protection assay to characterize the relative accumulation and abundance of individual adeno-associated virus type 2 (AAV) RNAs throughout the course of AAV-adenovirus coinfections and preinfections. We have demonstrated that there is a previously unrecognized temporal order to the appearance of AAV RNAs. First, unspliced P5-generated transcripts, which encode Rep78, were detectable prior to the significant accumulation of other AAV RNAs. Ultimately, as previously demonstrated, P19-generated products accumulated to levels greater than those generated from P5, and P40-generated transcripts predominated in the total RNA pool. Second, the percentage of each class of AAV RNA that was spliced increased during infection, and the degree of this increase was different for the P5/P19 products than for those generated by P40. At late times postcoinfection, approximately 90% of P40 products, but only approximately 50% of RNAs generated by P5 and P19, were seen to be spliced; thus, the AAV intron was removed to different final levels from these different RNA species. We have shown that each of the AAV RNAs is quite stable; the majority of each RNA species persisted 6 h after treatment with actinomycin D. Quantification of the accumulation of individual AAV RNAs, over intervals during which degradation was negligible, allowed us to infer that at late times during infection the relative strength of P5, P19, and P40 was approximately 1:3:18, respectively, consistent with the steady-state accumulated levels of the RNAs generated by each promoter. All AAV RNAs exited to the cytoplasm with similar efficiencies in the presence or absence of adenovirus; however, adenovirus coinfection appeared to stimulate total splicing of AAV RNAs and the relative use of the downstream intron acceptor. Our results confirm and extend previous observations concerning the appearance and processing of AAV-generated RNAs.
Figures









Similar articles
-
A Regulatory Element Near the 3' End of the Adeno-Associated Virus rep Gene Inhibits Adenovirus Replication in cis by Means of p40 Promoter-Associated Short Transcripts.J Virol. 2016 Mar 28;90(8):3981-93. doi: 10.1128/JVI.03120-15. Print 2016 Apr. J Virol. 2016. PMID: 26842470 Free PMC article.
-
Adeno-associated virus RNA transcription in vivo.Eur J Biochem. 1981 Dec;121(1):147-54. doi: 10.1111/j.1432-1033.1981.tb06443.x. Eur J Biochem. 1981. PMID: 6173214
-
Analysis of adeno-associated virus (AAV) wild-type and mutant Rep proteins for their abilities to negatively regulate AAV p5 and p19 mRNA levels.J Virol. 1994 May;68(5):2947-57. doi: 10.1128/JVI.68.5.2947-2957.1994. J Virol. 1994. PMID: 8151765 Free PMC article.
-
The Interplay between Adeno-Associated Virus and its Helper Viruses.Viruses. 2020 Jun 19;12(6):662. doi: 10.3390/v12060662. Viruses. 2020. PMID: 32575422 Free PMC article. Review.
-
Processing of adeno-associated virus RNA.Front Biosci. 2008 Jan 1;13:3101-15. doi: 10.2741/2912. Front Biosci. 2008. PMID: 17981780 Review.
Cited by
-
Efficient expression of the adeno-associated virus type 5 p41 capsid gene promoter in 293 cells does not require Rep.J Virol. 2006 Jul;80(13):6559-67. doi: 10.1128/JVI.00387-06. J Virol. 2006. PMID: 16775342 Free PMC article.
-
The adeno-associated virus type 5 small rep proteins expressed via internal translation initiation are functional.J Virol. 2013 Jan;87(1):296-303. doi: 10.1128/JVI.02547-12. Epub 2012 Oct 17. J Virol. 2013. PMID: 23077303 Free PMC article.
-
Upstream AP1- and CREB-binding sites confer high basal activity on the adeno-associated virus type 5 capsid gene promoter.J Virol. 2007 Mar;81(6):2605-13. doi: 10.1128/JVI.02313-06. Epub 2007 Jan 3. J Virol. 2007. PMID: 17202218 Free PMC article.
-
Characterization of the nonstructural proteins of the bocavirus minute virus of canines.J Virol. 2013 Jan;87(2):1098-104. doi: 10.1128/JVI.02627-12. Epub 2012 Nov 7. J Virol. 2013. PMID: 23135724 Free PMC article.
-
"D" matters in recombinant AAV DNA packaging.Mol Ther. 2021 Jun 2;29(6):1937-1939. doi: 10.1016/j.ymthe.2021.05.002. Epub 2021 May 19. Mol Ther. 2021. PMID: 34010593 Free PMC article. No abstract available.
References
-
- Berns K. Parvoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1017–1041.
-
- Berns K I, Giraud C. Biology of adeno-associated virus. Curr Top Microbiol Immunol. 1996;218:1–23. - PubMed
-
- Carter B J. Transcription. In: Ward D, Tattersall P, editors. Replication of mammalian proviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1978. pp. 33–52.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

