Heterogeneous spectrum of coreceptor usage among variants within a dualtropic human immunodeficiency virus type 1 primary-isolate quasispecies
- PMID: 11024154
- PMCID: PMC102064
- DOI: 10.1128/jvi.74.21.10229-10235.2000
Heterogeneous spectrum of coreceptor usage among variants within a dualtropic human immunodeficiency virus type 1 primary-isolate quasispecies
Abstract
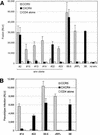
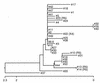
Human immunodeficiency virus type 1 (HIV-1) variants that use the coreceptor CCR5 for entry (R5; macrophage tropic) predominate in early infection, while variants that use CXCR4 emerge during disease progression. Some late-stage variants use CXCR4 alone (X4; T-cell tropic), while others use both CXCR4 and CCR5 (R5X4; dualtropic). It has been proposed that dualtropic R5X4 strains are intermediates in the evolution from R5 to X4, and we hypothesized that a dualtropic primary-isolate quasispecies might contain variants that represent the spectrum of coreceptor use in vivo. We generated a panel of 35 functional full-length env clones from the primary-isolate quasispecies of a dualtropic prototype strain, HIV-1 89.6(PI). Thirty of the functional env clones (86%) were R5X4, four (11%) were R5, and one (3%) was predominantly X4. V3 to V5 sequences did not reveal clustering by coreceptor usage, and no specific sequence motif or V3 charge pattern corresponded to coreceptor utilization. Complete sequencing of seven functionally divergent Env proteins revealed > or =98.7% homology and conservation of structurally important domains. Chimeras between the R5X4 89.6 prototype and an R5 variant indicated that multiple regions contributed to the use of CXCR4, while chimeras with the X4 variant implicated a single residue in V4 in CCR5 use. These results confirm, at the molecular level, both that dualtropic variants are a predominant component of late-stage syncytium-inducing isolates and that variants restricted to each coreceptor coexist with dualtropic species in vivo. Coreceptor-restricted minority variants may reflect residual R5 species from earlier in disease as well as emerging X4 variants.
Figures

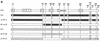
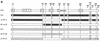
Similar articles
-
Concordant utilization of macrophage entry coreceptors by related variants within an HIV type 1 primary isolate viral swarm.AIDS Res Hum Retroviruses. 2001 Jul 1;17(10):957-63. doi: 10.1089/088922201750290078. AIDS Res Hum Retroviruses. 2001. PMID: 11461681
-
Differences in molecular evolution between switch (R5 to R5X4/X4-tropic) and non-switch (R5-tropic only) HIV-1 populations during infection.Infect Genet Evol. 2010 Apr;10(3):356-64. doi: 10.1016/j.meegid.2009.05.003. Epub 2009 May 14. Infect Genet Evol. 2010. PMID: 19446658
-
Purifying selection of CCR5-tropic human immunodeficiency virus type 1 variants in AIDS subjects that have developed syncytium-inducing, CXCR4-tropic viruses.J Gen Virol. 2006 May;87(Pt 5):1285-1294. doi: 10.1099/vir.0.81722-0. J Gen Virol. 2006. PMID: 16603531
-
Chemokine receptor utilization and macrophage signaling by human immunodeficiency virus type 1 gp120: Implications for neuropathogenesis.J Neurovirol. 2004;10 Suppl 1:91-6. doi: 10.1080/753312758. J Neurovirol. 2004. PMID: 14982745 Review.
-
HIV and the chemokine system: 10 years later.EMBO J. 2006 Feb 8;25(3):447-56. doi: 10.1038/sj.emboj.7600947. Epub 2006 Jan 26. EMBO J. 2006. PMID: 16437164 Free PMC article. Review.
Cited by
-
Clonal analysis of HIV-1 genotype and function associated with virologic failure in treatment-experienced persons receiving maraviroc: Results from the MOTIVATE phase 3 randomized, placebo-controlled trials.PLoS One. 2018 Dec 26;13(12):e0204099. doi: 10.1371/journal.pone.0204099. eCollection 2018. PLoS One. 2018. PMID: 30586365 Free PMC article. Clinical Trial.
-
Virus-host interactions in HIV pathogenesis: directions for therapy.Adv Dent Res. 2011 Apr;23(1):13-8. doi: 10.1177/0022034511398874. Adv Dent Res. 2011. PMID: 21441474 Free PMC article. Review.
-
Contrasting use of CCR5 structural determinants by R5 and R5X4 variants within a human immunodeficiency virus type 1 primary isolate quasispecies.J Virol. 2003 Nov;77(22):12057-66. doi: 10.1128/jvi.77.22.12057-12066.2003. J Virol. 2003. PMID: 14581542 Free PMC article.
-
Preferential suppression of CXCR4-specific strains of HIV-1 by antiviral therapy.J Clin Invest. 2001 Feb;107(4):431-8. doi: 10.1172/JCI11526. J Clin Invest. 2001. PMID: 11181642 Free PMC article.
-
Antigenic variation within the CD4 binding site of human immunodeficiency virus type 1 gp120: effects on chemokine receptor utilization.J Virol. 2001 Jun;75(12):5593-603. doi: 10.1128/JVI.75.12.5593-5603.2001. J Virol. 2001. PMID: 11356967 Free PMC article.
References
-
- Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. - PubMed
-
- Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

