Characterization of the cold stress-induced cyanobacterial DEAD-box protein CrhC as an RNA helicase
- PMID: 11024172
- PMCID: PMC110790
- DOI: 10.1093/nar/28.20.3926
Characterization of the cold stress-induced cyanobacterial DEAD-box protein CrhC as an RNA helicase
Abstract
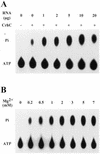
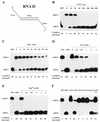
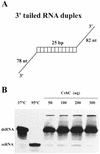
We have shown previously that CrhC is a unique member of the DEAD-box family of RNA helicases whose expression occurs specifically under conditions of cold stress. Here we show that recombinant His-tagged CrhC, purified from Escherichia coli, is an ATP-independent RNA binding protein possessing RNA-dependent ATPase activity which is stimulated most efficiently by rRNA and polysome preparations. RNA strand displacement assays indicate that CrhC possesses RNA unwinding activity that is adenosine nucleotide specific. Unwinding of partially duplexed RNA proceeds in the 5'-->3' but not the 3'-->5' direction using standard assay conditions. Immunoprecipitation and far-western analysis indicate that CrhC is a component of a multisubunit complex, interacting specifically with a 37 kDa polypeptide. We propose that CrhC unwinds cold-stabilized secondary structure in the 5'-UTR of RNA during cold stress.
Figures




Similar articles
-
Visualization of unwinding activity of duplex RNA by DbpA, a DEAD box helicase, at single-molecule resolution by atomic force microscopy.Proc Natl Acad Sci U S A. 2001 Apr 24;98(9):5007-12. doi: 10.1073/pnas.071372498. Epub 2001 Apr 10. Proc Natl Acad Sci U S A. 2001. PMID: 11296244 Free PMC article.
-
Regulation of cold shock-induced RNA helicase gene expression in the Cyanobacterium anabaena sp. strain PCC 7120.J Bacteriol. 2000 Mar;182(5):1251-6. doi: 10.1128/JB.182.5.1251-1256.2000. J Bacteriol. 2000. PMID: 10671444 Free PMC article.
-
Rearrangement of structured RNA via branch migration structures catalysed by the highly related DEAD-box proteins p68 and p72.Nucleic Acids Res. 2001 May 15;29(10):2088-96. doi: 10.1093/nar/29.10.2088. Nucleic Acids Res. 2001. PMID: 11353078 Free PMC article.
-
A new twist on RNA helicases: DExH/D box proteins as RNPases.Nat Struct Biol. 2001 Feb;8(2):113-6. doi: 10.1038/84091. Nat Struct Biol. 2001. PMID: 11175897 Review.
-
RNA helicases and remodeling proteins.Curr Opin Chem Biol. 2011 Oct;15(5):636-42. doi: 10.1016/j.cbpa.2011.07.019. Epub 2011 Aug 20. Curr Opin Chem Biol. 2011. PMID: 21862383 Free PMC article. Review.
Cited by
-
Effects of low temperature on tropical and temperate isolates of marine Synechococcus.ISME J. 2016 May;10(5):1252-63. doi: 10.1038/ismej.2015.179. Epub 2015 Oct 23. ISME J. 2016. PMID: 26495993 Free PMC article.
-
Liquid-Liquid Phase Separation of the DEAD-Box Cyanobacterial RNA Helicase Redox (CrhR) into Dynamic Membraneless Organelles in Synechocystis sp. Strain PCC 6803.Appl Environ Microbiol. 2023 Apr 26;89(4):e0001523. doi: 10.1128/aem.00015-23. Epub 2023 Mar 15. Appl Environ Microbiol. 2023. PMID: 36920190 Free PMC article.
-
RNA-binding strategies common to cold-shock domain- and RNA recognition motif-containing proteins.Nucleic Acids Res. 2001 Jun 1;29(11):2223-33. doi: 10.1093/nar/29.11.2223. Nucleic Acids Res. 2001. PMID: 11376140 Free PMC article.
-
Oligomerization and activity of the helicase domain of the tobacco mosaic virus 126- and 183-kilodalton replicase proteins.J Virol. 2003 Mar;77(6):3549-56. doi: 10.1128/jvi.77.6.3549-3556.2003. J Virol. 2003. PMID: 12610130 Free PMC article.
-
The Thermus thermophilus DEAD-box protein Hera is a general RNA binding protein and plays a key role in tRNA metabolism.RNA. 2020 Nov;26(11):1557-1574. doi: 10.1261/rna.075580.120. Epub 2020 Jul 15. RNA. 2020. PMID: 32669294 Free PMC article.
References
-
- Fuller-Pace F.V. (1994) Trends Cell Biol., 4, 271–274. - PubMed
-
- Luking A., Stahl,U. and Schmidt,U. (1998) Crit. Rev. Biochem. Mol. Biol., 33, 259–296. - PubMed
-
- Linder P., Lasko,P.F., Ashburner,M., Leroy,P., Nielsen,P.J., Nishi,K., Schnier,J. and Slonimsky,P.P. (1989) Nature, 337, 121–122. - PubMed
-
- Gorbalenya A.E. and Koonin,E.V. (1993). Curr. Opin. Struct. Biol., 3, 419–429.
-
- Pause A. and Sonenberg,N. (1993) Curr. Opin. Struct. Biol., 3, 953–959.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

