A heuristic graph comparison algorithm and its application to detect functionally related enzyme clusters
- PMID: 11024183
- PMCID: PMC110779
- DOI: 10.1093/nar/28.20.4021
A heuristic graph comparison algorithm and its application to detect functionally related enzyme clusters
Abstract
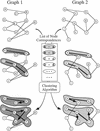
The availability of computerized knowledge on biochemical pathways in the KEGG database opens new opportunities for developing computational methods to characterize and understand higher level functions of complete genomes. Our approach is based on the concept of graphs; for example, the genome is a graph with genes as nodes and the pathway is another graph with gene products as nodes. We have developed a simple method for graph comparison to identify local similarities, termed correlated clusters, between two graphs, which allows gaps and mismatches of nodes and edges and is especially suitable for detecting biological features. The method was applied to a comparison of the complete genomes of 10 microorganisms and the KEGG metabolic pathways, which revealed, not surprisingly, a tendency for formation of correlated clusters called FRECs (functionally related enzyme clusters). However, this tendency varied considerably depending on the organism. The relative number of enzymes in FRECs was close to 50% for Bacillus subtilis and Escherichia coli, but was <10% for SYNECHOCYSTIS: and Saccharomyces cerevisiae. The FRECs collection is reorganized into a collection of ortholog group tables in KEGG, which represents conserved pathway motifs with the information about gene clusters in all the completely sequenced genomes.
Figures




References
-
- Kanehisa M. (1997) Trends Genet., 13, 375–376. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

