Automatic detection of conserved gene clusters in multiple genomes by graph comparison and P-quasi grouping
- PMID: 11024184
- PMCID: PMC110780
- DOI: 10.1093/nar/28.20.4029
Automatic detection of conserved gene clusters in multiple genomes by graph comparison and P-quasi grouping
Abstract
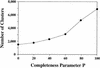
We previously reported two graph algorithms for analysis of genomic information: a graph comparison algorithm to detect locally similar regions called correlated clusters and an algorithm to find a graph feature called P-quasi complete linkage. Based on these algorithms we have developed an automatic procedure to detect conserved gene clusters and align orthologous gene orders in multiple genomes. In the first step, the graph comparison is applied to pairwise genome comparisons, where the genome is considered as a one-dimensionally connected graph with genes as its nodes, and correlated clusters of genes that share sequence similarities are identified. In the next step, the P-quasi complete linkage analysis is applied to grouping of related clusters and conserved gene clusters in multiple genomes are identified. In the last step, orthologous relations of genes are established among each conserved cluster. We analyzed 17 completely sequenced microbial genomes and obtained 2313 clusters when the completeness parameter P: was 40%. About one quarter contained at least two genes that appeared in the metabolic and regulatory pathways in the KEGG database. This collection of conserved gene clusters is used to refine and augment ortholog group tables in KEGG and also to define ortholog identifiers as an extension of EC numbers.
Figures




References
-
- Tatusov R.L., Mushegian,A.R., Bork,P., Brown,N.P., Hayes,W.S., Borodovsky,M., Rudd,K.E. and Koonin,V. (1996) Curr. Biol., 6, 279–291. - PubMed
-
- Watanabe H., Mori,H., Itoh,T. and Gojobori,T. (1997) J. Mol. Evol., 44, S57–S64. - PubMed
-
- Siefert J.L., Martin,K.A., Abdi,F., Widger,W.R. and Fox,G.E. (1997) J. Mol. Evol., 45, 467–472. - PubMed
-
- Tamames J., Casari,G., Ouzounis,C. and Valencia,A. (1997) J. Mol. Evol., 44, 66–73. - PubMed
-
- Dandekar T., Snel,B., Huynen,M. and Bork,P. (1998) Trends Biochem. Sci., 23, 324–328. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

