Solid phase DNA amplification: characterisation of primer attachment and amplification mechanisms
- PMID: 11024189
- PMCID: PMC110803
- DOI: 10.1093/nar/28.20.e87
Solid phase DNA amplification: characterisation of primer attachment and amplification mechanisms
Abstract
Different chemical methods used to attach oligonucleotides by their 5'-end on a glass surface were tested in the framework of solid phase PCR where surface-bound instead of freely-diffusing primers are used to amplify DNA. Each method was first evaluated for its capacity to provide a high surface coverage of oligonucleotides essentially attached via a 5'-specific linkage that satisfyingly withstands PCR conditions and leaves the 3'-ends available for DNA polymerase activity. The best results were obtained with 5'-thiol-modified oligonucleotides attached to amino-silanised glass slides using a heterobifunctional cross-linker reagent. It was then demonstrated that the primers bound to the glass surface using the optimal chemistry can be involved in attaching and amplifying DNA molecules present in the reaction mix in the absence of freely-diffusing primers. Two distinct amplification processes called interfacial and surface amplification have been observed and characterised. The newly synthesised DNA can be detected and quantified by radioactive and fluorescent hybridisation assays. These new surface amplification processes are seen as an interesting approach for attachment of DNA molecules by their 5'-end on a solid support and can be used as an alternative route for producing DNA chips for genomic studies.
Figures

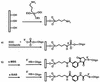




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

