Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis
- PMID: 11025658
- PMCID: PMC1255923
- DOI: 10.1038/35036309
Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis
Abstract
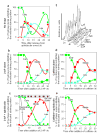
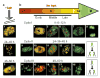
Here we show that exposure of aphidicolin-arrested Chinese hamster ovary (CHO) cells to the protein-kinase inhibitors 2-aminopurine or caffeine results in initiation of replication at successively later-replicating chromosomal domains, loss of the capacity to synthesize DNA at earlier-replicating sites, release of Mcm2 proteins from chromatin, and redistribution of PCNA and RPA from early- to late-replicating domains in the absence of detectable elongation of replication forks. These results provide evidence that, under conditions of replicational stress, checkpoint controls not only prevent further initiation but may also be required to actively maintain the integrity of stalled replication complexes.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

