Functional characterization of nuclear localization signals in yeast Sm proteins
- PMID: 11027265
- PMCID: PMC86405
- DOI: 10.1128/MCB.20.21.7943-7954.2000
Functional characterization of nuclear localization signals in yeast Sm proteins
Abstract
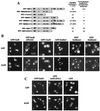
In mammals, nuclear localization of U-snRNP particles requires the snRNA hypermethylated cap structure and the Sm core complex. The nature of the signal located within the Sm core proteins is still unknown, both in humans and yeast. Close examination of the sequences of the yeast SmB, SmD1, and SmD3 carboxyl-terminal domains reveals the presence of basic regions that are reminiscent of nuclear localization signals (NLSs). Fluorescence microscopy studies using green fluorescent protein (GFP)-fusion proteins indicate that both yeast SmB and SmD1 basic amino acid stretches exhibit nuclear localization properties. Accordingly, deletions or mutations in the NLS-like motifs of SmB and SmD1 dramatically reduce nuclear fluorescence of the GFP-Sm mutant fusion alleles. Phenotypic analyses indicate that the NLS-like motifs of SmB and SmD1 are functionally redundant: each NLS-like motif can be deleted without affecting yeast viability whereas a simultaneous deletion of both NLS-like motifs is lethal. Taken together, these findings suggest that, in the doughnut-like structure formed by the Sm core complex, the carboxyl-terminal extensions of Sm proteins may form an evolutionarily conserved basic amino acid-rich protuberance that functions as a nuclear localization determinant.
Figures




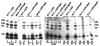


References
-
- Bordonné R, Banroques J, Abelson J, Guthrie C. Domains of yeast U4 spliceosomal RNA required for PRP4 protein binding, snRNP-snRNP interactions, and pre-mRNA splicing in vivo. Genes Dev. 1990;4:1185–1196. - PubMed
-
- Bordonné R, Tarassov I. The yeast SME1 gene encodes the homologue of the human E core protein. Gene. 1996;176:111–117. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
