DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links
- PMID: 11027296
- PMCID: PMC86436
- DOI: 10.1128/MCB.20.21.8283-8289.2000
DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links
Abstract
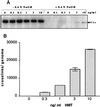
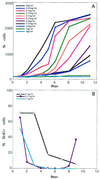
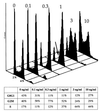
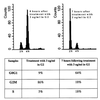
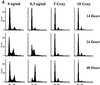
Following introduction of DNA interstrand cross-links (ICLs), mammalian cells display chromosome breakage or cell cycle delay with a 4N DNA content. To further understand the nature of the delay, previously described as a G(2)/M arrest, we developed a protocol to generate ICLs during specific intervals of the cell cycle. Synchronous populations of G(1), S, and G(2) cells were treated with photoactivated 4'-hydroxymethyl-4,5',8-trimethylpsoralen (HMT) and scored for normal passage into mitosis. In contrast to what was found for ionizing radiation, ICLs introduced during G(2) did not result in a G(2)/M arrest, mitotic arrest, or chromosome breakage. Rather, subsequent passage through S phase was required to trigger both chromosome breakage and arrest in the next cell cycle. Similarly, ICLs introduced during G(1) did not cause a G(1)/S arrest. We conclude that DNA replication is required to elicit the cellular responses of cell cycle arrest and genomic instability after psoralen-induced ICLs. In primary human fibroblasts, the 4N DNA content cell cycle arrest triggered by ICLs was long lasting but reversible. Kinetic analysis suggested that these cells could remove up to approximately 2,500 ICLs/genome at an average rate of 11 ICLs/genome/h.
Figures



References
-
- Auerbach A D, Wolman S R. Susceptibility of Fanconi's anaemia fibroblasts to chromosome damage by carcinogens. Nature. 1976;261:494–496. - PubMed
-
- Bempong M A, Trower E C. Sensitivity of rat testes to inhibitors of nucleic acid synthesis. III. The inheritance of mitomycin C-induced structural rearrangements of chromosomes. J Hered. 1975;66:285–289. - PubMed
-
- Cheng S, Van Houten B, Gamper H B, Sancar A, Hearst J E. Use of psoralen-modified oligonucleotides to trap three-stranded RecA-DNA complexes and repair of these cross-linked complexes by ABC excinuclease. J Biol Chem. 1988;263:15110–15117. - PubMed
-
- de Andrade H H, Marques E K, Schenberg A C, Henriques J A. The PSO4 gene is responsible for an error-prone recombinational DNA repair pathway in Saccharomyces cerevisiae. Mol Gen Genet. 1989;217:419–426. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
