The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways
- PMID: 11029042
- PMCID: PMC14998
- DOI: 10.1091/mbc.11.10.3365
The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways
Abstract
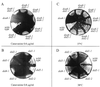
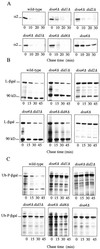
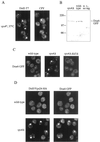
The Saccharomyces cerevisiae DOA4 gene encodes a deubiquitinating enzyme that is required for rapid degradation of ubiquitin-proteasome pathway substrates. Both genetic and biochemical data suggest that Doa4 acts in this pathway by facilitating ubiquitin recycling from ubiquitinated intermediates targeted to the proteasome. Here we describe the isolation of 12 spontaneous extragenic suppressors of the doa4-1 mutation; these involve seven different genes, six of which were cloned. Surprisingly, all of the cloned DID (Doa4-independent degradation) genes encode components of the vacuolar protein-sorting (Vps) pathway. In particular, all are class E Vps factors, which function in the maturation of a late endosome/prevacuolar compartment into multivesicular bodies that then fuse with the vacuole. Four of the six Did proteins are structurally related, suggesting an overlap in function. In wild-type and several vps strains, Doa4-green fluorescent protein displays a cytoplasmic/nuclear distribution. However, in cells lacking the Vps4/Did6 ATPase, a large fraction of Doa4-green fluorescent protein, like several other Vps factors, concentrates at the late endosome-like class E compartment adjacent to the vacuole. These results suggest an unanticipated connection between protein deubiquitination and endomembrane protein trafficking in which Doa4 acts at the late endosome/prevacuolar compartment to recover ubiquitin from ubiquitinated membrane proteins en route to the vacuole.
Figures







References
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1989.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

