Role for the silencing protein Dot1 in meiotic checkpoint control
- PMID: 11029058
- PMCID: PMC15018
- DOI: 10.1091/mbc.11.10.3601
Role for the silencing protein Dot1 in meiotic checkpoint control
Abstract
During the meiotic cell cycle, a surveillance mechanism called the "pachytene checkpoint" ensures proper chromosome segregation by preventing meiotic progression when recombination and chromosome synapsis are defective. The silencing protein Dot1 (also known as Pch1) is required for checkpoint-mediated pachytene arrest of the zip1 and dmc1 mutants of Saccharomyces cerevisiae. In the absence of DOT1, the zip1 and dmc1 mutants inappropriately progress through meiosis, generating inviable meiotic products. Other components of the pachytene checkpoint include the nucleolar protein Pch2 and the heterochromatin component Sir2. In dot1, disruption of the checkpoint correlates with the loss of concentration of Pch2 and Sir2 in the nucleolus. In addition to its checkpoint function, Dot1 blocks the repair of meiotic double-strand breaks by a Rad54-dependent pathway of recombination between sister chromatids. In vegetative cells, mutation of DOT1 results in delocalization of Sir3 from telomeres, accounting for the impaired telomeric silencing in dot1.
Figures

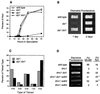
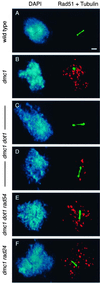
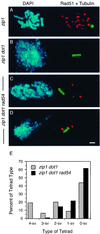
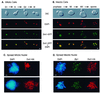
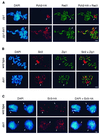
References
-
- Ahmed S, Hodgkin J. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature. 2000;403:159–164. - PubMed
-
- Bailis JM, Roeder GS. The pachytene checkpoint. Trends Genet. 2000a;16:395–403. - PubMed
-
- Bailis JM, Roeder GS. Pachytene exit controlled by reversal of Mek1-dependent phosphorylation. Cell. 2000b;101:211–221. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

