Role of TnrA in nitrogen source-dependent repression of Bacillus subtilis glutamate synthase gene expression
- PMID: 11029411
- PMCID: PMC94725
- DOI: 10.1128/JB.182.21.5939-5947.2000
Role of TnrA in nitrogen source-dependent repression of Bacillus subtilis glutamate synthase gene expression
Abstract
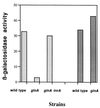
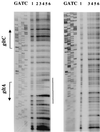
Synthesis of glutamate, the cell's major donor of nitrogen groups and principal anion, occupies a significant fraction of bacterial metabolism. In Bacillus subtilis, the gltAB operon, encoding glutamate synthase, requires a specific positive regulator, GltC, for its expression. In addition, the gltAB operon was shown to be repressed by TnrA, a regulator of several other genes of nitrogen metabolism and active under conditions of ammonium (nitrogen) limitation. TnrA was found to bind directly to a site immediately downstream of the gltAB promoter. As is true for other genes, the activity of TnrA at the gltAB promoter was antagonized by glutamine synthetase under certain growth conditions.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

