RNA polymerases from Bacillus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro
- PMID: 11029421
- PMCID: PMC94735
- DOI: 10.1128/JB.182.21.6027-6035.2000
RNA polymerases from Bacillus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro
Erratum in
- J Bacteriol 2001 Feb;183(4):1504
Abstract
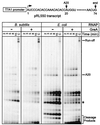
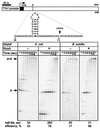
Adaptation of bacterial cells to diverse habitats relies on the ability of RNA polymerase to respond to various regulatory signals. Some of these signals are conserved throughout evolution, whereas others are species specific. In this study we present a comprehensive comparative analysis of RNA polymerases from two distantly related bacterial species, Escherichia coli and Bacillus subtilis, using a panel of in vitro transcription assays. We found substantial species-specific differences in the ability of these enzymes to escape from the promoter and to recognize certain types of elongation signals. Both enzymes responded similarly to other pause and termination signals and to the general E. coli elongation factors NusA and GreA. We also demonstrate that, although promoter recognition depends largely on the sigma subunit, promoter discrimination exhibited in species-specific fashion by both RNA polymerases resides in the core enzyme. We hypothesize that differences in signal recognition are due to the changes in contacts made between the beta and beta' subunits and the downstream DNA duplex.
Figures





References
-
- Anthony L, Artsimovitch I, Svetlov V, Landick R, Burgess R R. Rapid purification of the His(6)-tagged Bacillus subtilis core RNA polymerase. Protein Expr Purif. 2000;19:350–354. - PubMed
-
- Babitzke P. Regulation of tryptophan biosynthesis: Trp-ing the TRAP or how Bacillus subtilis reinvented the wheel. Mol Microbiol. 1997;26:1–9. - PubMed
-
- Bartlett M S, Gaal T, Ross W, Gourse R L. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J Mol Biol. 1998;279:331–345. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

