Mapping of the Cryptococcus neoformans MATalpha locus: presence of mating type-specific mitogen-activated protein kinase cascade homologs
- PMID: 11029445
- PMCID: PMC94759
- DOI: 10.1128/JB.182.21.6222-6227.2000
Mapping of the Cryptococcus neoformans MATalpha locus: presence of mating type-specific mitogen-activated protein kinase cascade homologs
Abstract
In this study we investigated the relationship between the MATalpha locus of Cryptococcus neoformans and several MATalpha-specific mitogen-activated protein (MAP) kinase signal transduction cascade genes, including STE12alpha, STE11alpha, and STE20alpha. To resolve the location of the genes, we screened a cosmid library of the MATalpha strain B-4500 (JEC21), which was chosen for the C. neoformans genome project. We isolated several overlapping cosmids spanning a region of about 71 kb covering the entire MATalpha locus. It was found that STE12alpha, STE11alpha, and STE20alpha are imbedded within the locus rather than closely linked to the locus. Furthermore, three copies of MFalpha, the mating type alpha-pheromone gene, a MATalpha-specific myosin gene, and a pheromone receptor (CPRalpha) were identified within the locus. We created a physical map, based on the restriction enzyme BamHI, and identified both borders of the MATalpha locus. The MATalpha locus of C. neoformans is approximately 50 kb in size and is one of the largest mating type loci reported among fungi with a one-locus, two-allele mating system.
Figures

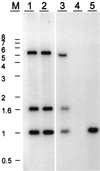
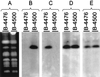

References
-
- Bölker M, Urban M, Kahmann R. The a mating type locus of U. maydis specifies cell signaling components. Cell. 1992;68:441–450. - PubMed
-
- Chaturvedi S, Rodeghier B, Fan J, McClelland C M, Wickes B L, Chaturvedi V. Direct PCR of Cryptococcus neoformans MATα and MATa pheromones to determine mating type, ploidy, and variety: a tool for epidemiological and molecular pathogenesis studies. J Clin Microbiol. 2000;38:2007–2009. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

