Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system
- PMID: 11032813
- PMCID: PMC314003
- DOI: 10.1093/emboj/19.20.5460
Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system
Abstract
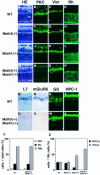
Whereas vertebrate achaete-scute complex (as-c) and atonal (ato) homologs are required for neurogenesis, their neuronal determination activities in the central nervous system (CNS) are not yet supported by loss-of-function studies, probably because of genetic redundancy. Here, to address this problem, we generated mice double mutant for the as-c homolog Mash1 and the ato homolog Math3. Whereas in Mash1 or Math3 single mutants neurogenesis is only weakly affected, in the double mutants tectal neurons, two longitudinal columns of hindbrain neurons and retinal bipolar cells were missing and, instead, those cells that normally differentiate into neurons adopted the glial fate. These results indicated that Mash1 and Math3 direct neuronal versus glial fate determination in the CNS and raised the possibility that downregulation of these bHLH genes is one of the mechanisms to initiate gliogenesis.
Figures

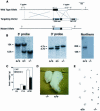
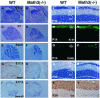

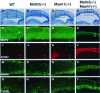
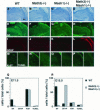
References
-
- Akazawa C., Sasai,Y., Nakanishi,S. and Kageyama,R. (1992) Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J. Biol. Chem., 267, 21879–21885. - PubMed
-
- Akazawa C., Ishibashi,M., Shimizu,C., Nakanishi,S. and Kageyama,R. (1995) A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J. Biol. Chem., 270, 8730–8738. - PubMed
-
- Anderson D.J. (1999) Lineages and transcription factors in the specification of vertebrate primary sensory neurons. Curr. Opin. Neurobiol., 9, 517–524. - PubMed
-
- Anderson D.J. and Jan,Y.N. (1997) The determination of the neuronal phenotype. In Cowan,W.M. (ed.), Molecular and Cellular Approaches to Neural Development. Oxford University Press, New York, NY, pp. 26–63.
-
- Ben-Arie N., Bellen,H.J, Armstrong,D.L., McCall,A.E., Gordadze,P.R., Guo,Q., Matzuk,M.M. and Zoghbi,H.Y. (1997) Math1 is essential for genesis of cerebellar granule neurons. Nature, 390, 169–172. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

