Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination
- PMID: 11032822
- PMCID: PMC313999
- DOI: 10.1093/emboj/19.20.5552
Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination
Abstract
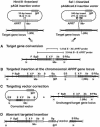
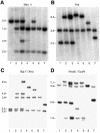
The XpF/Ercc1 structure-specific endonuclease performs the 5' incision in nucleotide excision repair and is the apparent mammalian counterpart of the Rad1/Rad10 endonuclease from Saccharomyces cerevisiae. In yeast, Rad1/Rad10 endonuclease also functions in mitotic recombination. To determine whether XpF/Ercc1 endonuclease has a similar role in mitotic recombination, we targeted the APRT locus in Chinese hamster ovary ERCC1(+) and ERCC1(-) cell lines with insertion vectors having long or short terminal non-homologies flanking each side of a double-strand break. No substantial differences were evident in overall recombination frequencies, in contrast to results from targeting experiments in yeast. However, profound differences were observed in types of APRT(+) recombinants recovered from ERCC1(-) cells using targeting vectors with long terminal non-homologies-almost complete ablation of gap repair and single-reciprocal exchange events, and generation of a new class of aberrant insertion/deletion recombinants absent in ERCC1(+) cells. These results represent the first demonstration of a requirement for ERCC1 in targeted homologous recombination in mammalian cells, specifically in removal of long non-homologous tails from invading homologous strands.
Figures


References
-
- Aboussekhra A. and Wood,R.D. (1994) Repair of UV-damaged DNA by mammalian cells and Saccharomyces cerevisiae. Curr. Opin. Genet. Dev., 4, 212–220. - PubMed
-
- Aboussekhra A. et al. (1995) Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell, 80, 859–868. - PubMed
-
- Adair G.M., Scheerer,J.B., Brotherman,A., McConville,S., Wilson,J.H. and Nairn,R.S. (1998) Targeted recombination at the Chinese hamster APRT locus using insertion versus replacement vectors. Somat. Cell Mol. Genet., 24, 91–105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

