Antigenic variation of Anaplasma marginale by expression of MSP2 mosaics
- PMID: 11035716
- PMCID: PMC97690
- DOI: 10.1128/IAI.68.11.6133-6138.2000
Antigenic variation of Anaplasma marginale by expression of MSP2 mosaics
Abstract
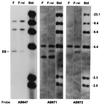
Anaplasma marginale is a tick-borne pathogen, one of several closely related ehrlichial organisms that cause disease in animals and humans. These Ehrlichia species have complex life cycles that require, in addition to replication and development within the tick vector, evasion of the immune system in order to persist in the mammalian reservoir host. This complexity requires efficient use of the small ehrlichial genome. A. marginale and related ehrlichiae express immunoprotective, variable outer membrane proteins that have similar structures and are encoded by polymorphic multigene families. We show here that the major outer membrane protein of A. marginale, MSP2, is encoded on a polycistronic mRNA. The genomic expression site for this mRNA is polymorphic and encodes numerous amino acid sequence variants in bloodstream populations of A. marginale. A potential mechanism for persistence is segmental gene conversion of the expression site to link hypervariable msp2 sequences to the promoter and polycistron.
Figures




References
-
- Alleman A R, Kamper S M, Viseshakul N, Barbet A F. Analysis of the Anaplasma marginale genome by pulsed-field electrophoresis. J Gen Microbiol. 1993;139:2439–2444. - PubMed
-
- Andersson S G, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. - PubMed
-
- Barbet A F, Kamper S M. The importance of mosaic genes to trypanosome survival. Parasitol Today. 1993;9:63–66. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

