Coupled two-way clustering analysis of gene microarray data
- PMID: 11035779
- PMCID: PMC17297
- DOI: 10.1073/pnas.210134797
Coupled two-way clustering analysis of gene microarray data
Abstract
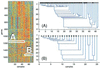
We present a coupled two-way clustering approach to gene microarray data analysis. The main idea is to identify subsets of the genes and samples, such that when one of these is used to cluster the other, stable and significant partitions emerge. The search for such subsets is a computationally complex task. We present an algorithm, based on iterative clustering, that performs such a search. This analysis is especially suitable for gene microarray data, where the contributions of a variety of biological mechanisms to the gene expression levels are entangled in a large body of experimental data. The method was applied to two gene microarray data sets, on colon cancer and leukemia. By identifying relevant subsets of the data and focusing on them we were able to discover partitions and correlations that were masked and hidden when the full dataset was used in the analysis. Some of these partitions have clear biological interpretation; others can serve to identify possible directions for future research.
Figures

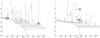
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

