The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway
- PMID: 11038174
- PMCID: PMC2192639
- DOI: 10.1083/jcb.151.2.263
The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway
Abstract
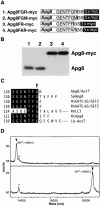
Autophagy and the Cvt pathway are examples of nonclassical vesicular transport from the cytoplasm to the vacuole via double-membrane vesicles. Apg8/Aut7, which plays an important role in the formation of such vesicles, tends to bind to membranes in spite of its hydrophilic nature. We show here that the nature of the association of Apg8 with membranes changes depending on a series of modifications of the protein itself. First, the carboxy-terminal Arg residue of newly synthesized Apg8 is removed by Apg4/Aut2, a novel cysteine protease, and a Gly residue becomes the carboxy-terminal residue of the protein that is now designated Apg8FG. Subsequently, Apg8FG forms a conjugate with an unidentified molecule "X" and thereby binds tightly to membranes. This modification requires the carboxy-terminal Gly residue of Apg8FG and Apg7, a ubiquitin E1-like enzyme. Finally, the adduct Apg8FG-X is reversed to soluble or loosely membrane-bound Apg8FG by cleavage by Apg4. The mode of action of Apg4, which cleaves both newly synthesized Apg8 and modified Apg8FG, resembles that of deubiquitinating enzymes. A reaction similar to ubiquitination is probably involved in the second modification. The reversible modification of Apg8 appears to be coupled to the membrane dynamics of autophagy and the Cvt pathway.
Figures






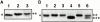
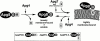
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

