Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division
- PMID: 11038182
- PMCID: PMC2192646
- DOI: 10.1083/jcb.151.2.353
Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division
Abstract
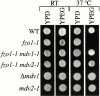
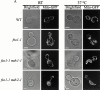
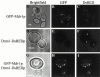
Mitochondrial fission is mediated by the dynamin-related GTPase, Dnm1p, which assembles on the mitochondrial outer membrane into punctate structures associated with sites of membrane constriction and fission. We have identified additional nuclear genes required for mitochondrial fission, termed MDV (for mitochondrial division). MDV1 encodes a predicted soluble protein, containing a coiled-coil motif and seven COOH-terminal WD repeats. Genetic and two-hybrid analyses indicate that Mdv1p interacts with Dnm1p to mediate mitochondrial fission. In addition, Mdv1p colocalizes with Dnm1p in fission-mediating punctate structures on the mitochondrial outer membrane. Whereas localization of Mdv1p to these structures requires Dnm1p, localization of Mdv1p to mitochondrial membranes does not. This indicates that Mdv1p possesses a Dnm1p-independent mitochondrial targeting signal. Dnm1p-independent targeting of Mdv1p to mitochondria requires MDV2. Our data indicate that MDV2 also functions separately to regulate the assembly of Dnm1p into punctate structures. In contrast, Mdv1p is not required for the assembly of Dnm1p, but Dnm1p-containing punctate structures lacking Mdv1p are not able to complete division. Our studies suggest that mitochondrial fission is a multi-step process in which Mdv2p regulates the assembly of Dnm1p into punctate structures and together with Mdv1p functions later during fission to facilitate Dnm1p-dependent mitochondrial membrane constriction and/or division.
Figures



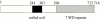





Comment in
-
A mitochondrial division apparatus takes shape.J Cell Biol. 2000 Oct 16;151(2):F1-4. doi: 10.1083/jcb.151.2.f1. J Cell Biol. 2000. PMID: 11038192 Free PMC article. No abstract available.
References
-
- Cziepluch C., Kordes E., Pujol A., Jauniax J.-C. Sequencing analysis of a 40.2 kb fragment of yeast chromosome X reveals 19 open reading frames including URA2 (5′ end), TRK1, PBS2, SPT10, GCD14, RPE1, PH086, NCA3, ASF1, CCT7, CZF3, two tRNA genes, three remnant delta elements and a Ty4 transposon. Yeast. 1996;12:1471–1474. - PubMed
-
- Gietz R.D., Schiestl R.H. Transforming yeast with DNA. Methods Mol. Cell. Biol. 1994;5:255–269.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

