The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition
- PMID: 11038188
- PMCID: PMC2192652
- DOI: 10.1083/jcb.151.2.425
The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition
Abstract
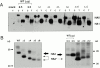
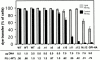
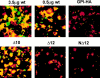
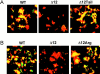
Glycosylphosphatidylinositol-anchored influenza hemagglutinin (GPI-HA) mediates hemifusion, whereas chimeras with foreign transmembrane (TM) domains mediate full fusion. A possible explanation for these observations is that the TM domain must be a critical length in order for HA to promote full fusion. To test this hypothesis, we analyzed biochemical properties and fusion phenotypes of HA with alterations in its 27-amino acid TM domain. Our mutants included sequential 2-amino acid (Delta2-Delta14) and an 11-amino acid deletion from the COOH-terminal end, deletions of 6 or 8 amino acids from the NH(2)-terminal and middle regions, and a deletion of 12 amino acids from the NH(2)-terminal end of the TM domain. We also made several point mutations in the TM domain. All of the mutants except Delta14 were expressed at the cell surface and displayed biochemical properties virtually identical to wild-type HA. All the mutants that were expressed at the cell surface promoted full fusion, with the notable exception of deletions of >10 amino acids. A mutant in which 11 amino acids were deleted was severely impaired in promoting full fusion. Mutants in which 12 amino acids were deleted (from either end) mediated only hemifusion. Hence, a TM domain of 17 amino acids is needed to efficiently promote full fusion. Addition of either the hydrophilic HA cytoplasmic tail sequence or a single arginine to Delta12 HA, the hemifusion mutant that terminates with 15 (hydrophobic) amino acids of the HA TM domain, restored full fusion activity. Our data support a model in which the TM domain must span the bilayer to promote full fusion.
Figures







Comment in
-
HIV-1 membrane fusion: targets of opportunity.J Cell Biol. 2000 Oct 16;151(2):F9-14. doi: 10.1083/jcb.151.2.f9. J Cell Biol. 2000. PMID: 11038194 Free PMC article. No abstract available.
Similar articles
-
Amino acid sequence requirements of the transmembrane and cytoplasmic domains of influenza virus hemagglutinin for viable membrane fusion.Mol Biol Cell. 1999 Jun;10(6):1821-36. doi: 10.1091/mbc.10.6.1821. Mol Biol Cell. 1999. PMID: 10359599 Free PMC article.
-
Meta-stability of the hemifusion intermediate induced by glycosylphosphatidylinositol-anchored influenza hemagglutinin.Biophys J. 1997 Nov;73(5):2280-91. doi: 10.1016/S0006-3495(97)78260-2. Biophys J. 1997. PMID: 9370425 Free PMC article.
-
Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin-induced hemifusion to full fusion.J Cell Biol. 1997 Mar 10;136(5):995-1005. doi: 10.1083/jcb.136.5.995. J Cell Biol. 1997. PMID: 9060465 Free PMC article.
-
Membrane fusion mechanisms: the influenza hemagglutinin paradigm and its implications for intracellular fusion.Traffic. 2000 Aug;1(8):598-604. doi: 10.1034/j.1600-0854.2000.010803.x. Traffic. 2000. PMID: 11208147 Review.
-
Structural and functional specificity of Influenza virus haemagglutinin and paramyxovirus fusion protein anchoring peptides.Virus Res. 2017 Jan 2;227:183-199. doi: 10.1016/j.virusres.2016.09.014. Epub 2016 Oct 20. Virus Res. 2017. PMID: 27773768 Review.
Cited by
-
A novel function for the sperm adhesion protein IZUMO1 in cell-cell fusion.J Cell Biol. 2023 Feb 6;222(2):e202207147. doi: 10.1083/jcb.202207147. Epub 2022 Nov 17. J Cell Biol. 2023. PMID: 36394541 Free PMC article.
-
The conserved glycine-rich segment linking the N-terminal fusion peptide to the coiled coil of human T-cell leukemia virus type 1 transmembrane glycoprotein gp21 is a determinant of membrane fusion function.J Virol. 2005 Apr;79(7):4533-9. doi: 10.1128/JVI.79.7.4533-4539.2005. J Virol. 2005. PMID: 15767455 Free PMC article.
-
Evidences for the existence of intermolecular disulfide-bonded oligomers in the H3 hemagglutinins expressed in insect cells.Virus Genes. 2014 Apr;48(2):304-11. doi: 10.1007/s11262-013-1021-0. Epub 2013 Dec 3. Virus Genes. 2014. PMID: 24297311
-
Conformation and Trimer Association of the Transmembrane Domain of the Parainfluenza Virus Fusion Protein in Lipid Bilayers from Solid-State NMR: Insights into the Sequence Determinants of Trimer Structure and Fusion Activity.J Mol Biol. 2018 Mar 2;430(5):695-709. doi: 10.1016/j.jmb.2018.01.002. Epub 2018 Jan 10. J Mol Biol. 2018. PMID: 29330069 Free PMC article.
-
Capture and release of partially zipped trans-SNARE complexes on intact organelles.J Cell Biol. 2009 May 4;185(3):535-49. doi: 10.1083/jcb.200811082. J Cell Biol. 2009. PMID: 19414611 Free PMC article.
References
-
- Bullough P.A., Hughson F.M., Skehel J.J., Wiley D.C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. - PubMed
-
- Carr C.M., Kim P.S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. - PubMed
-
- Chernomordik L.V., Melikyan G.B., Chizmadzhev Y.A. Biomembrane fusiona new concept derived from model studies using two interacting planar lipid bilayers. Biochim. Biophys. Acta. 1987;906:309–352. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

