High-resolution mapping and isolation of a yeast artificial chromosome contig containing fw2.2: a major fruit weight quantitative trait locus in tomato
- PMID: 11038534
- PMCID: PMC26434
- DOI: 10.1073/pnas.93.26.15503
High-resolution mapping and isolation of a yeast artificial chromosome contig containing fw2.2: a major fruit weight quantitative trait locus in tomato
Abstract
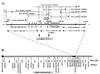
A high-resolution physical and genetic map of a major fruit weight quantitative trait locus (QTL), fw2.2, has been constructed for a region of tomato chromosome 2. Using an F(2) nearly isogenic line mapping population (3472 individuals) derived from Lycopersicon esculentum (domesticated tomato) x Lycopersicon pennellii (wild tomato), fw2.2 has been placed near TG91 and TG167, which have an interval distance of 0.13 +/- 0.03 centimorgan. The physical distance between TG91 and TG167 was estimated to be </= 150 kb by pulsed-field gel electrophoresis of tomato DNA. A physical contig composed of six yeast artificial chromosomes (YACs) and encompassing fw2.2 was isolated. No rearrangements or chimerisms were detected within the YAC contig based on restriction fragment length polymorphism analysis using YAC-end sequences and anchored molecular markers from the high-resolution map. Based on genetic recombination events, fw2.2 could be narrowed down to a region less than 150 kb between molecular markers TG91 and HSF24 and included within two YACs: YAC264 (210 kb) and YAC355 (300 kb). This marks the first time, to our knowledge, that a QTL has been mapped with such precision and delimited to a segment of cloned DNA. The fact that the phenotypic effect of the fw2.2 QTL can be mapped to a small interval suggests that the action of this QTL is likely due to a single gene. The development of the high-resolution genetic map, in combination with the physical YAC contig, suggests that the gene responsible for this QTL and other QTLs in plants can be isolated using a positional cloning strategy. The cloning of fw2.2 will likely lead to a better understanding of the molecular biology of fruit development and to the genetic engineering of fruit size characteristics.
Figures

References
-
- Johanssen W. Elemente der exakten Erblichkeitsllehre. Jena, Germany: Fischer; 1909.
-
- Nilsson-Ehle H. Lunds Univ Arsskr Avd 2. 1909;5:1–122.
-
- Paterson A H, Lander E S, Hewitt J D, Peterson S, Lincoln S E, Tanksley S D. Nature (London) 1988;335:721–726. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

