DNA sequence evidence for the segmental allotetraploid origin of maize
- PMID: 11038553
- PMCID: PMC21240
- DOI: 10.1073/pnas.94.13.6809
DNA sequence evidence for the segmental allotetraploid origin of maize
Abstract
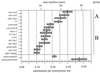
It has long been suspected that maize is the product of an historical tetraploid event. Several observations support this possibility, including the fact that the maize genome contains duplicated chromosomal segments with colinear gene arrangements. Some of the genes from these duplicated segments have been sequenced. In this study, we examine the pattern of sequence divergence among 14 pairs of duplicated genes. We compare the pattern of divergence to patterns predicted by four models of the evolution of the maize genome-autotetraploidy, genomic allotetraploidy, segmental allotetraploidy, and multiple segmental duplications. Our analyses indicate that coalescent times for duplicated sequences fall into two distinct groups, corresponding to roughly 20.5 and 11.4 million years. This observation strongly discounts the possibility that the maize genome is the product of a genomic allotetraploid event, and it is also difficult to reconcile with either autotetraploidy or multiple independent segmental duplications. However, the presence of two (and only two) coalescent times is predicted by the segmental allotetraploid model. If the maize genome is the product of a segmental allotetraploid event, as these data suggest, then its two diploid progenitors diverged roughly 20.5 million years ago (Mya), and the allotetraploid event probably occurred approximately 11.4 Mya. Comparison of maize and sorghum sequences suggests that one of the two ancestral diploids shares a more recent common ancestor with sorghum than it does with the other ancestral diploid.
Figures


References
-
- Celarier R P. Rhodora. 1956;58:135–143.
-
- Anderson E. Chron Bot. 1945;9:88–92.
-
- Garber E D. Univ Calif Pub Bot. 1950;23:283–362.
-
- Rhoades M M. Am Nat. 1951;85:105–110.
-
- Wendel J F, Stuber C W, Goodman M M, Beckett J B. J Hered. 1989;80:218–228. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

