Limitations of clonality analysis of B cell proliferations using CDR3 polymerase chain reaction
- PMID: 11040942
- PMCID: PMC1186969
- DOI: 10.1136/mp.53.4.194
Limitations of clonality analysis of B cell proliferations using CDR3 polymerase chain reaction
Abstract
Background/aims: Detection of clonal immunoglobulin heavy chain (IgH) rearrangements by the polymerase chain reaction (PCR) is an attractive alternative to Southern blotting in lymphoma diagnostics. However, the advantages and limitations of PCR in clonality analysis are still not fully appreciated. In this study, clonality was analysed by means of PCR, focusing in particular on the sample size requirements when studying extremely small samples of polyclonal and monoclonal lesions.
Materials/methods: High resolution complementarity determining region 3 (CDR3) PCR was used to investigate the minimum number of cells and the amount of tissue required for the detection of a polyclonal population, both for fresh cells and formalin fixed, paraffin wax embedded tissue. Subsequently, frozen and paraffin wax embedded samples of 76 B cell lymphoproliferative disorders, 43 of which were tested by means of Southern blotting, were analysed to establish the sensitivity of this assay. These specimens included 12 chronic lymphocytic leukaemias (CLLs), nine mantle cell lymphomas (MCLs), 10 follicular lymphomas (FLs), and 45 mucosa associated lymphoid tissue (MALT) lymphomas. The specificity was tested on reactive lymph nodes (n = 19), tonsils (n = 4), peripheral blood lymphocyte fractions (n = 4), and biopsies with gastritis (n = 21).
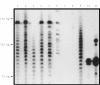
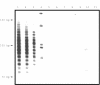
Results: In reactive tissue, 20 ng of high molecular weight DNA derived from 6.5-9 x 10(3) B cells was sufficient to obtain a polyclonal PCR result. With smaller amounts "pseudoclonality" could be induced. When using paraffin wax blocks, undiluted DNA isolated from tonsillar tissue of at least 1 mm2 was necessary to obtain a polyclonal pattern. The sensitivity required to detect clonality in paraffin wax embedded and frozen tissue by PCR for FL (40% and 60%, respectively) was lower than that for MALT lymphomas (60% and 86%, respectively), CLL (78% and 89%, respectively), and MCL (88% and 100%, respectively). PCR specificity was 96% and 100% for frozen and paraffin wax embedded tissue, respectively.
Conclusion: The minimum amount of template for CDR3 PCR is approximately 20 ng of high molecular weight DNA or 1 mm3 of B cell rich paraffin wax embedded normal tonsillar tissue, but care has to be taken to avoid pseudoclonality when low numbers of B cells are present. Duplicate or triplicate tests should be performed to avoid misinterpretation. The specificity of the PCR assay is almost 100%, whereas sensitivity depends on a combination of factors, such as lymphoma type and tissue fixation. Because frozen samples yield better results, obtaining fresh material for the PCR assay is recommended, especially when analysing FL and MALT lymphomas.
Figures
Similar articles
-
Increased sensitivity of B-cell clonality analysis in formalin-fixed and paraffin-embedded B-cell lymphoma samples using an enzyme blend with both 5'-->3' DNA polymerase and 3'-->5' exonuclease activity.Virchows Arch. 2003 Nov;443(5):643-8. doi: 10.1007/s00428-003-0880-5. Epub 2003 Aug 21. Virchows Arch. 2003. PMID: 12937979
-
Detection of immunoglobulin gene rearrangement of B cell non-Hodgkin's lymphomas and leukemias in fresh, unfixed and formalin-fixed, paraffin-embedded tissue by polymerase chain reaction.Lab Invest. 1993 Jun;68(6):746-57. Lab Invest. 1993. PMID: 8515660
-
Detection of clonal B cells in microdissected reactive lymphoproliferations: possible diagnostic pitfalls in PCR analysis of immunoglobulin heavy chain gene rearrangement.Mol Pathol. 1999 Apr;52(2):104-10. doi: 10.1136/mp.52.2.104. Mol Pathol. 1999. PMID: 10474690 Free PMC article.
-
Intravascular (angiotropic) large cell lymphoma: determination of monoclonality by polymerase chain reaction on paraffin-embedded tissues.Mod Pathol. 1994 Jun;7(5):593-8. Mod Pathol. 1994. PMID: 7524070 Review.
-
[Clonality analysis of B-cell lymphoproliferative disorders by means of immunoglobulin heavy chain polymerase reaction].Orv Hetil. 1996 Sep 8;137(36):1963-7. Orv Hetil. 1996. PMID: 8927349 Review. Hungarian.
Cited by
-
Optimization of PCR amplification for B- and T-cell clonality analysis on formalin-fixed and paraffin-embedded samples.Pathol Oncol Res. 2007;13(3):209-14. doi: 10.1007/BF02893501. Epub 2007 Oct 7. Pathol Oncol Res. 2007. PMID: 17922050
-
Inconclusive flow cytometric surface light chain results; can cytoplasmic light chains, Bcl-2 expression and PCR clonality analysis improve accuracy of cytological diagnoses in B-cell lymphomas?Diagn Pathol. 2015 Oct 20;10:191. doi: 10.1186/s13000-015-0427-5. Diagn Pathol. 2015. PMID: 26482649 Free PMC article.
-
Splitting bone marrow trephines into frozen and fixed fragments allows parallel histological and molecular detection of B cell malignant infiltrates.J Clin Pathol. 2006 Oct;59(10):1111-3. doi: 10.1136/jcp.2005.030379. J Clin Pathol. 2006. PMID: 17021140 Free PMC article.
-
High-dose methotrexate is beneficial in parenchymal brain masses of uncertain origin suspicious for primary CNS lymphoma.Neuro Oncol. 2007 Apr;9(2):96-102. doi: 10.1215/15228517-2006-037. Epub 2007 Feb 14. Neuro Oncol. 2007. PMID: 17301290 Free PMC article.
-
Clonality analysis of lymphoid proliferations using the BIOMED-2 clonality assays: a single institution experience.Radiol Oncol. 2014 Apr 25;48(2):155-62. doi: 10.2478/raon-2013-0072. eCollection 2014 Jun. Radiol Oncol. 2014. PMID: 24991205 Free PMC article.
References
-
- van Dongen JJ, Wolvers-Tettero IL. Analysis of immunoglobulin and T cell receptor genes. Part I: basic and technical aspects. Clin Chim Acta 1991;198:1–91. - PubMed
-
- Trainor KJ, Brisco MJ, Story CJ, et al. Monoclonality in B-lymphoproliferative disorders detected at the DNA level. Blood 1990;75:2220–2. - PubMed
-
- Arnold A, Cossman J, Bakhshi A, et al. Immunoglobulin-gene rearrangements as unique clonal markers in human lymphoid neoplasms. N Engl J Med 1983;309:1593–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
