Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species from a bone marrow transplant recipient
- PMID: 11040945
- PMCID: PMC1186972
- DOI: 10.1136/mp.53.4.211
Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species from a bone marrow transplant recipient
Abstract
Aims: To ascertain the clinical relevance of a strain of Enterobacteriaceae isolated from the stool of a bone marrow transplant recipient with diarrhoea. The isolate could not be identified to the genus level by conventional phenotypic methods and required 16S ribosomal RNA (rRNA) gene sequencing for full identification.
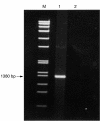
Methods: The isolate was investigated phenotypically by standard biochemical methods using conventional biochemical tests and two commercially available systems, the Vitek (GNI+) and API (20E) systems. Genotypically, the 16S bacterial rRNA gene was amplified by the polymerase chain reaction (PCR) and sequenced. The sequence of the PCR product was compared with known 16S rRNA gene sequences in the GenBank database by multiple sequence alignment.
Results: Conventional biochemical tests did not reveal a pattern resembling any known member of the Enterobacteriaceae family. The isolate was identified as Salmonella arizonae (73%) and Escherichia coli (76%) by the Vitek (GNI+) and API (20E) systems, respectively. 16S rRNA sequencing showed that there was only one base difference between the isolate and E coli K-12, but 48 and 47 base differences between the isolate and S typhimurium (NCTC 8391) and S typhi (St111), respectively, showing that it was an E coli strain. The patient did not require any specific treatment and the diarrhoea subsided spontaneously.
Conclusions: 16S rRNA gene sequencing was useful in ascertaining the clinical relevance of the strain of Enterobacteriaceae isolated from the stool of the bone marrow transplant recipient with diarrhoea.
Figures

References
-
- Tomb JF, White O, Kerlavage AR, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 1997;388:539–47. - PubMed
-
- Olsen GJ, Woese CR. Ribosomal RNA: a key to phylogeny. FASEB J 1993;7:113–23. - PubMed
-
- Relman DA, Loutit JS, Schmidt TM, et al. The agent of bacillary angiomatosis. N Engl J Med 1990;323:1573–80. - PubMed
-
- Relman DA, Schmidt TM, MacDermott RP, et al. Identification of the uncultured bacillus of Whipple's disease. N Engl J Med 1992;327:293–301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
