Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems
- PMID: 11041893
- PMCID: PMC149136
- DOI: 10.1105/tpc.12.10.2001
Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems
Abstract
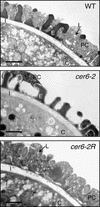
Very long chain lipids contribute to the hydrophobic cuticle on the surface of all land plants and are an essential component of the extracellular pollen coat in the Brassicaceae. Mutations in Arabidopsis CER genes eliminate very long chain lipids from the cuticle surface and, in some cases, from the pollen coat. In Arabidopsis, the loss of pollen coat lipids can disrupt interactions with the stigma, inhibiting pollen hydration and causing sterility. We have positionally cloned CER6 and demonstrate that a wild-type copy complements the cer6-2 defect. In addition, we have identified a fertile, intragenic suppressor, cer6-2R, that partially restores pollen coat lipids but does not rescue the stem wax defect, suggesting an intriguing difference in the requirements for CER6 activity on stems and the pollen coat. Importantly, analysis of this suppressor demonstrates that low amounts of very long chain lipids are sufficient for pollen hydration and germination. The predicted CER6 amino acid sequence resembles that of fatty acid-condensing enzymes, consistent with its role in the production of epicuticular and pollen coat lipids >28 carbons long. DNA sequence analysis revealed the nature of the cer6-1, cer6-2, and cer6-2R mutations, and segregation analysis showed that CER6 is identical to CUT1, a cDNA previously mapped to a different chromosome arm. Instead, we have determined that a new gene, CER60, with a high degree of nucleotide and amino acid similarity to CER6, resides at the original CUT1 locus.
Figures




References
-
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. - PubMed
-
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–143. - PubMed
-
- Hannoufa, A., McNevin, J.P., and Lemieux, B. (1993). Epicuticular wax of Arabidopsis thaliana eceriferum (cer) mutants. Phytochemistry 33, 851–855. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

