Sequence diversity and large-scale typing of SNPs in the human apolipoprotein E gene
- PMID: 11042151
- PMCID: PMC310963
- DOI: 10.1101/gr.146900
Sequence diversity and large-scale typing of SNPs in the human apolipoprotein E gene
Abstract
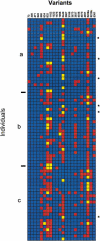
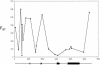
A common strategy for genotyping large samples begins with the characterization of human single nucleotide polymorphisms (SNPs) by sequencing candidate regions in a small sample for SNP discovery. This is usually followed by typing in a large sample those sites observed to vary in a smaller sample. We present results from a systematic investigation of variation at the human apolipoprotein E locus (APOE), as well as the evaluation of the two-tiered sampling strategy based on these data. We sequenced 5.5 kb spanning the entire APOE genomic region in a core sample of 72 individuals, including 24 each of African-Americans from Jackson, Mississippi; European-Americans from Rochester, Minnesota; and Europeans from North Karelia, Finland. This sequence survey detected 21 SNPs and 1 multiallelic indel, 14 of which had not been previously reported. Alleles varied in relative frequency among the populations, and 10 sites were polymorphic in only a single population sample. Oligonucleotide ligation assays (OLA) were developed for 20 of these sites (omitting the indel and a closely-linked SNP). These were then scored in 2179 individuals sampled from the same three populations (n = 843, 884, and 452, respectively). Relative allele frequencies were generally consistent with estimates from the core sample, although variation was found in some populations in the larger sample at SNPs that were monomorphic in the corresponding smaller core sample. Site variation in the larger samples showed no systematic deviation from Hardy-Weinberg expectation. The large OLA sample clearly showed that variation in many, but not all, of OLA-typed SNPs is significantly correlated with the classical protein-coding variants, implying that there may be important substructure within the classical epsilon 2, epsilon 3, and epsilon 4 alleles. Comparison of the levels and patterns of polymorphism in the core samples with those estimated for the OLA-typed samples shows how nucleotide diversity is underestimated when only a subset of sites are typed and underscores the importance of adequate population sampling at the polymorphism discovery stage. [The sequence data described in this paper have been submitted to the GenBank data library under accession no. AF261279.]
Figures

References
-
- Artiga MJ, Bullido MJ, Frank A, Sastre I, Recuero M, Garcia MA, Lendon CL, Han SW, Morris JC, Vazquez J, et al. Risk for Alzheimer's disease correlates with transcriptional activity of the APOEgene. Hum Mol Genet. 1998a;7:1887–1892. - PubMed
-
- Artiga MJ, Bullido MJ, Sastre I, Recuero M, Gracia MA, Aldudo J, Vazquez J, Valdivieso F. Allelic polymorphisms in the transcriptional regulatory region of apolipoprotein E gene. FEBS Lett. 1998b;421:105–108. - PubMed
-
- Bullido MJ, Artiga MJ, Recuero M, Sastre I, Garcia MA, Aldudo J, Lendon C, Han SW, Morris JC, Frank A, et al. A polymorphism in the regulatory region of APOE associated with risk for Alzheimer's dementia. Nat Genet. 1998;18:69–71. - PubMed
-
- Collins FS, Guyer MS, Chakravarti A. Variations on a theme: Cataloging human DNA sequence variation. Science. 1997;278:1580–1581. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
