Dependence of adenovirus infectivity on length of the fiber shaft domain
- PMID: 11044071
- PMCID: PMC110901
- DOI: 10.1128/jvi.74.22.10274-10286.2000
Dependence of adenovirus infectivity on length of the fiber shaft domain
Abstract
One of the objectives in adenovirus (Ad) vector development is to target gene delivery to specific cell types. Major attention has been given to modification of the Ad fiber knob, which is thought to determine virus tropism. However, among the human Ad serotypes with different tissue tropisms, not only the knob but also the length of the fiber shaft domain varies significantly. In this study we attempted to delineate the role of fiber length in coxsackievirus-adenovirus receptor (CAR)- and non-CAR-mediated infection. A series of Ad serotype 5 (Ad5) capsid-based vectors containing long or short fibers with knob domains derived from Ad5, Ad9, or Ad35 was constructed and tested in adsorption, internalization, and transduction studies. For Ad5 or Ad9 knob-possessing vectors, a long-shafted fiber was critical for efficient adsorption/internalization and transduction of CAR/alphav integrin-expressing cells. Ad5 capids containing short CAR-recognizing fibers were affected in cell adsorption and infection. In contrast, for the chimeric vectors possessing Ad35 knobs, which enter cells by a CAR/alphav integrin-independent pathway, fiber shaft length had no significant influence on binding or infectibility on tested cells. The weak attachment of short-shafted Ad5 or Ad9 knob-possessing vectors seems to be causally associated with a charge-dependent repulsion between Ad5 capsid and acidic cell surface proteins. The differences between short- and long-shafted vectors in attachment or infection were abrogated by preincubation of cells with polycations. This study demonstrates that the fiber-CAR interaction is not the sole determinant for tropism of Ad vectors containing chimeric fibers. CAR- and alphav integrin-mediated infections are influenced by other factors, including the length of the fiber shaft.
Figures


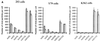
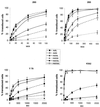
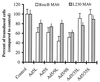
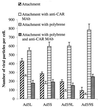
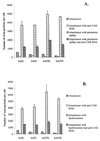
 ) or be internalized into (□) cells as described in Materials and Methods. Cells were then washed, and the number of viral particles bound per cell was determined. The data were obtained from two to four independent experiments performed in triplicate. (B) To analyze the level of Ad attachment, equal amounts of indicated Ad vectors at an MOI of 8,000 genomes per cell were mixed with 3.5 × 105 293 cells in 100 μl of adhesion buffer and then allowed to attach for 1 h on ice. Then virus-containing medium was removed; cells were washed twice with ice-cold PBS and resuspended in 100 μl of adhesion buffer. Then 100 μl of lysis buffer was added to the cells, and DNA was extracted as described earlier (61) (Virus attachment). To estimate the amount of Ad loaded per sample, the same volume of corresponding Ad as used in the attachment study was mixed with 3.5 × 105 293 cells in 100 μl of adhesion buffer. Immediately after mixing, 100 μl of lysis buffer was added to cells, and DNA was extracted (Virus load). After purification, DNA concentration was measured spectrophotometrically, and 1 μg of each sample was applied on the agarose gel. Shown are the ethidium bromide-stained 1% agarose gel before blotting, demonstrating that similar amounts of genomic DNA were loaded (top), and the result of Southern blot hybridization of the transferred DNA with a 32P-labeled 8-kb HindIII fragment of the Ad5 genome, corresponding to the E2 region (bottom). The conditions of DNA transfer and hybridization were described earlier (61). M, molecular weight marker. Arrows indicate the Ad DNA (bottom) or mixture of Ad and cellular DNA (top).
) or be internalized into (□) cells as described in Materials and Methods. Cells were then washed, and the number of viral particles bound per cell was determined. The data were obtained from two to four independent experiments performed in triplicate. (B) To analyze the level of Ad attachment, equal amounts of indicated Ad vectors at an MOI of 8,000 genomes per cell were mixed with 3.5 × 105 293 cells in 100 μl of adhesion buffer and then allowed to attach for 1 h on ice. Then virus-containing medium was removed; cells were washed twice with ice-cold PBS and resuspended in 100 μl of adhesion buffer. Then 100 μl of lysis buffer was added to the cells, and DNA was extracted as described earlier (61) (Virus attachment). To estimate the amount of Ad loaded per sample, the same volume of corresponding Ad as used in the attachment study was mixed with 3.5 × 105 293 cells in 100 μl of adhesion buffer. Immediately after mixing, 100 μl of lysis buffer was added to cells, and DNA was extracted (Virus load). After purification, DNA concentration was measured spectrophotometrically, and 1 μg of each sample was applied on the agarose gel. Shown are the ethidium bromide-stained 1% agarose gel before blotting, demonstrating that similar amounts of genomic DNA were loaded (top), and the result of Southern blot hybridization of the transferred DNA with a 32P-labeled 8-kb HindIII fragment of the Ad5 genome, corresponding to the E2 region (bottom). The conditions of DNA transfer and hybridization were described earlier (61). M, molecular weight marker. Arrows indicate the Ad DNA (bottom) or mixture of Ad and cellular DNA (top).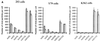
 ) or be internalized into (□) cells as described in Materials and Methods. Cells were then washed, and the number of viral particles bound per cell was determined. The data were obtained from two to four independent experiments performed in triplicate. (B) To analyze the level of Ad attachment, equal amounts of indicated Ad vectors at an MOI of 8,000 genomes per cell were mixed with 3.5 × 105 293 cells in 100 μl of adhesion buffer and then allowed to attach for 1 h on ice. Then virus-containing medium was removed; cells were washed twice with ice-cold PBS and resuspended in 100 μl of adhesion buffer. Then 100 μl of lysis buffer was added to the cells, and DNA was extracted as described earlier (61) (Virus attachment). To estimate the amount of Ad loaded per sample, the same volume of corresponding Ad as used in the attachment study was mixed with 3.5 × 105 293 cells in 100 μl of adhesion buffer. Immediately after mixing, 100 μl of lysis buffer was added to cells, and DNA was extracted (Virus load). After purification, DNA concentration was measured spectrophotometrically, and 1 μg of each sample was applied on the agarose gel. Shown are the ethidium bromide-stained 1% agarose gel before blotting, demonstrating that similar amounts of genomic DNA were loaded (top), and the result of Southern blot hybridization of the transferred DNA with a 32P-labeled 8-kb HindIII fragment of the Ad5 genome, corresponding to the E2 region (bottom). The conditions of DNA transfer and hybridization were described earlier (61). M, molecular weight marker. Arrows indicate the Ad DNA (bottom) or mixture of Ad and cellular DNA (top).
) or be internalized into (□) cells as described in Materials and Methods. Cells were then washed, and the number of viral particles bound per cell was determined. The data were obtained from two to four independent experiments performed in triplicate. (B) To analyze the level of Ad attachment, equal amounts of indicated Ad vectors at an MOI of 8,000 genomes per cell were mixed with 3.5 × 105 293 cells in 100 μl of adhesion buffer and then allowed to attach for 1 h on ice. Then virus-containing medium was removed; cells were washed twice with ice-cold PBS and resuspended in 100 μl of adhesion buffer. Then 100 μl of lysis buffer was added to the cells, and DNA was extracted as described earlier (61) (Virus attachment). To estimate the amount of Ad loaded per sample, the same volume of corresponding Ad as used in the attachment study was mixed with 3.5 × 105 293 cells in 100 μl of adhesion buffer. Immediately after mixing, 100 μl of lysis buffer was added to cells, and DNA was extracted (Virus load). After purification, DNA concentration was measured spectrophotometrically, and 1 μg of each sample was applied on the agarose gel. Shown are the ethidium bromide-stained 1% agarose gel before blotting, demonstrating that similar amounts of genomic DNA were loaded (top), and the result of Southern blot hybridization of the transferred DNA with a 32P-labeled 8-kb HindIII fragment of the Ad5 genome, corresponding to the E2 region (bottom). The conditions of DNA transfer and hybridization were described earlier (61). M, molecular weight marker. Arrows indicate the Ad DNA (bottom) or mixture of Ad and cellular DNA (top).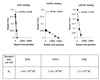

References
-
- Arcasoy S M, Latoche J, Gondor M, Watkins S C, Henderson R A, Hughey R, Finn O J, Pilewski J M. MUC1 and other sialoglycoconjugates inhibit adenovirus-mediated gene transfer to epithelial cells. Am J Respir Cell Mol Biol. 1997;17:422–435. - PubMed
-
- Arnberg N, Mei Y, Wadell G. Fiber genes of adenoviruses with tropism for the eye and the genital tract. Virology. 1997;227:239–244. - PubMed
-
- Bailey A, Mautner V. Phylogenetic relationships among adenovirus serotypes. Virology. 1994;205:438–452. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

