Generation of subgenomic RNA directed by a satellite RNA associated with bamboo mosaic potexvirus: analyses of potexvirus subgenomic RNA promoter
- PMID: 11044078
- PMCID: PMC110908
- DOI: 10.1128/jvi.74.22.10341-10348.2000
Generation of subgenomic RNA directed by a satellite RNA associated with bamboo mosaic potexvirus: analyses of potexvirus subgenomic RNA promoter
Abstract
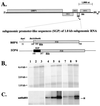
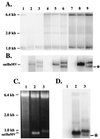
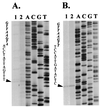
Satellite RNA of bamboo mosaic potexvirus (satBaMV), a single-stranded positive-sense RNA encoding a nonstructural protein of 20 kDa (P20), depends on bamboo mosaic potexvirus (BaMV) for replication and encapsidation. A full-length cDNA clone of satBaMV was used to examine the sequences required for the synthesis of potexvirus subgenomic RNAs (sgRNAs). Subgenomic promoter-like sequences (SGPs), 107 nucleotides (nt) upstream from the capsid protein (CP) gene of BaMV-V, were inserted upstream of the start codon of the P20 gene of satBaMV. Insertion of SGPs gave rise to the synthesis of sgRNA of satBaMV in protoplasts of Nicotiana benthamiana and leaves of Chenopodium quinoa when coinoculated with BaMV-V genomic RNA. Moreover, both the satBaMV cassette and its sgRNA were encapsidated. From analysis of the SGPs by deletion mutation, we concluded that an SGP contains one core promoterlike sequence (nt -30 through +16), two upstream enhancers (nt -59 through -31 and -91 through -60), and one downstream enhancer (nt +17 through +52), when the transcription initiation site is taken as +1. Site-directed mutagenesis and compensatory mutation to disrupt and restore potential base pairing in the core promoter-like sequence suggest that the stem-loop structure is important for the function of SGP in vivo. Likewise, the insertion of a putative SGP of the BaMV open reading frame 2 gene or a heterologous SGP of potato virus X resulted in generation of an sgRNA. The satBaMV cassette should be a useful tool to gain insight into sequences required for the synthesis of potexvirus sgRNAs.
Figures




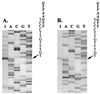
Similar articles
-
The Functional Roles of the Cis-acting Elements in Bamboo mosaic virus RNA Genome.Front Microbiol. 2017 Apr 13;8:645. doi: 10.3389/fmicb.2017.00645. eCollection 2017. Front Microbiol. 2017. PMID: 28450857 Free PMC article. Review.
-
Structural and mutational analyses of cis-acting sequences in the 5'-untranslated region of satellite RNA of bamboo mosaic potexvirus.Virology. 2003 Jun 20;311(1):229-39. doi: 10.1016/s0042-6822(03)00178-8. Virology. 2003. PMID: 12832220
-
The open reading frame of bamboo mosaic potexvirus satellite RNA is not essential for its replication and can be replaced with a bacterial gene.Proc Natl Acad Sci U S A. 1996 Apr 2;93(7):3138-42. doi: 10.1073/pnas.93.7.3138. Proc Natl Acad Sci U S A. 1996. PMID: 8610182 Free PMC article.
-
Crucial role of the 5' conserved structure of bamboo mosaic virus satellite RNA in downregulation of helper viral RNA replication.J Virol. 2006 Mar;80(5):2566-74. doi: 10.1128/JVI.80.5.2566-2574.2006. J Virol. 2006. PMID: 16474162 Free PMC article.
-
Interfering Satellite RNAs of Bamboo mosaic virus.Front Microbiol. 2017 May 4;8:787. doi: 10.3389/fmicb.2017.00787. eCollection 2017. Front Microbiol. 2017. PMID: 28522996 Free PMC article. Review.
Cited by
-
NbPsbO1 Interacts Specifically with the Bamboo Mosaic Virus (BaMV) Subgenomic RNA (sgRNA) Promoter and Is Required for Efficient BaMV sgRNA Transcription.J Virol. 2021 Sep 27;95(20):e0083121. doi: 10.1128/JVI.00831-21. Epub 2021 Aug 11. J Virol. 2021. PMID: 34379502 Free PMC article.
-
The Replicase Protein of Potato Virus X Is Able to Recognize and Trans-Replicate Its RNA Component.Viruses. 2024 Oct 15;16(10):1611. doi: 10.3390/v16101611. Viruses. 2024. PMID: 39459944 Free PMC article.
-
Development of expression vectors based on pepino mosaic virus.Plant Methods. 2011 Mar 11;7:6. doi: 10.1186/1746-4811-7-6. Plant Methods. 2011. PMID: 21396092 Free PMC article.
-
Production of Human IFNγ Protein in Nicotiana benthamiana Plant through an Enhanced Expression System Based on Bamboo mosaic Virus.Viruses. 2019 Jun 3;11(6):509. doi: 10.3390/v11060509. Viruses. 2019. PMID: 31163694 Free PMC article.
-
The Functional Roles of the Cis-acting Elements in Bamboo mosaic virus RNA Genome.Front Microbiol. 2017 Apr 13;8:645. doi: 10.3389/fmicb.2017.00645. eCollection 2017. Front Microbiol. 2017. PMID: 28450857 Free PMC article. Review.
References
-
- Balmori E, Gilmer D, Richards K, Guilley H, Jonard G. Mapping the promoter for subgenomic RNA synthesis on beet necrotic yellow vein virus RNA 3. Biochimie. 1993;75:517–521. - PubMed
-
- Beck D L, Guilford D M V, Andersen M T, Forster R L S. Triple gene block proteins of white clover mosaic potexvirus are required for transport. Virology. 1991;183:695–702. - PubMed
-
- Chang R C, Chen J C, Shaw J F. Facile purification of highly active recombinant Staphylococcus hyicus lipase fragment and characterization of a putative Lid region. Biochem Biophy Res Commun. 1996;228:774–779. - PubMed
-
- Chapman S, Kavanagh T A, Baulcombe D C. Potato virus X as a vector for gene expression in plants. Plant J. 1992;2:549–557. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

