Toward a comprehensive phylogeny for mammalian and avian herpesviruses
- PMID: 11044084
- PMCID: PMC110914
- DOI: 10.1128/jvi.74.22.10401-10406.2000
Toward a comprehensive phylogeny for mammalian and avian herpesviruses
Abstract
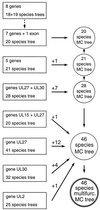
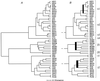
With the aim of deriving a definitive phylogenetic tree for as many mammalian and avian herpesvirus species as possible, alignments were made of amino acid sequences from eight conserved and ubiquitously present genes of herpesviruses, with 48 virus species each represented by at least one gene. Phylogenetic trees for both single-gene and concatenated alignments were evaluated thoroughly by maximum-likelihood methods, with each of the three herpesvirus subfamilies (the Alpha-, Beta-, and Gammaherpesvirinae) examined independently. Composite trees were constructed starting with the top-scoring tree based on the broadest set of genes and supplemented by addition of virus species from trees based on narrower gene sets, to give finally a 46-species tree; branching order for three regions within the tree remained unresolved. Sublineages of the Alpha- and Betaherpesvirinae showed extensive cospeciation with host lineages by criteria of congruence in branching patterns and consistency in extent of divergence. The Gammaherpesvirinae presented a more complex picture, with both higher and lower substitution rates in different sublineages. The final tree obtained represents the most detailed view to date of phylogenetic relationships in any family of large-genome viruses.
Figures



References
-
- Adachi J, Hasegawa M. The MOLPHY 2.2 package. Tokyo, Japan: Institute of Statistical Mathematics; 1994.
-
- Davison A J. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. - PubMed
-
- Davison A J, Sauerbier E, Dolan A, Addison C, McKinnell R G. Genomic studies of the Lucké tumor herpesvirus (RaHV-1) J Cancer Res Clin Oncol. 1999;125:232–238. - PubMed
-
- Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

