Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding
- PMID: 11044087
- PMCID: PMC110917
- DOI: 10.1128/jvi.74.22.10430-10437.2000
Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding
Abstract
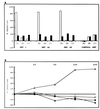
The hepatitis C virus (HCV) internal ribosome entry site (IRES) is a highly structured RNA element that directs cap-independent translation of the viral polyprotein. Morpholino antisense oligonucleotides directed towards stem loop IIId drastically reduced HCV IRES activity. Mutagenesis studies of this region showed that the GGG triplet (nucleotides 266 through 268) of the hexanucleotide apical loop of stem loop IIId is essential for IRES activity both in vitro and in vivo. Sequence comparison showed that apical loop nucleotides (UUGGGU) were absolutely conserved across HCV genotypes and the GGG triplet was strongly conserved among related Flavivirus and Pestivirus nontranslated regions. Chimeric IRES elements with IIId derived from GB virus B (GBV-B) in the context of the HCV IRES possess translational activity. Mutations within the IIId stem loop that abolish IRES activity also affect the RNA structure in RNase T(1)-probing studies, demonstrating the importance of correct RNA folding to IRES function.
Figures
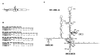






Similar articles
-
Mutational analysis of the GB virus B internal ribosome entry site.J Virol. 2000 Jan;74(2):773-83. doi: 10.1128/jvi.74.2.773-783.2000. J Virol. 2000. PMID: 10623739 Free PMC article.
-
Down-regulation of the internal ribosome entry site (IRES)-mediated translation of the hepatitis C virus: critical role of binding of the stem-loop IIId domain of IRES and the viral core protein.Virology. 2006 Feb 20;345(2):434-45. doi: 10.1016/j.virol.2005.10.013. Epub 2005 Nov 17. Virology. 2006. PMID: 16297950
-
A phylogenetically conserved stem-loop structure at the 5' border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation.J Virol. 1999 Feb;73(2):1165-74. doi: 10.1128/JVI.73.2.1165-1174.1999. J Virol. 1999. PMID: 9882318 Free PMC article.
-
Hepatitis C IRES: translating translation into a therapeutic target.Curr Opin Mol Ther. 2001 Jun;3(3):278-87. Curr Opin Mol Ther. 2001. PMID: 11497352 Review.
-
Oligonucleotide-based strategies to inhibit human hepatitis C virus.Oligonucleotides. 2003;13(6):539-48. doi: 10.1089/154545703322860834. Oligonucleotides. 2003. PMID: 15025918 Review.
Cited by
-
Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation.RNA. 2006 Feb;12(2):223-34. doi: 10.1261/rna.2153206. Epub 2005 Dec 22. RNA. 2006. PMID: 16373480 Free PMC article.
-
Structural features of the Seneca Valley virus internal ribosome entry site (IRES) element: a picornavirus with a pestivirus-like IRES.J Virol. 2011 May;85(9):4452-61. doi: 10.1128/JVI.01107-10. Epub 2011 Feb 16. J Virol. 2011. PMID: 21325406 Free PMC article.
-
Using Morpholinos to Control Gene Expression.Curr Protoc Nucleic Acid Chem. 2017 Mar 2;68(1):4.30.1-4.30.29. doi: 10.1002/cpnc.21. Curr Protoc Nucleic Acid Chem. 2017. PMID: 28252184 Free PMC article.
-
LOOP IIId of the HCV IRES is essential for the structural rearrangement of the 40S-HCV IRES complex.Nucleic Acids Res. 2016 Feb 18;44(3):1309-25. doi: 10.1093/nar/gkv1325. Epub 2015 Nov 30. Nucleic Acids Res. 2016. PMID: 26626152 Free PMC article.
-
Structure of the three-way helical junction of the hepatitis C virus IRES element.RNA. 2010 Aug;16(8):1597-609. doi: 10.1261/rna.2158410. Epub 2010 Jun 25. RNA. 2010. PMID: 20581129 Free PMC article.
References
-
- Alt M, Renz R, Hofschneider P H, Paumgartner G, Caselmann W H. Specific inhibition of hepatitis C viral gene expression by antisense phosphorothioate oligodeoxynucleotides. Hepatology. 1995;22:707–717. - PubMed
-
- Antivirals, Inc. Technical bulletin no. 2. Corvallis, Oreg: Antivirals, Inc.; 1997.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

