Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis
- PMID: 11046155
- PMCID: PMC102165
- DOI: 10.1128/MCB.20.22.8602-8612.2000
Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis
Abstract
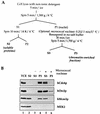
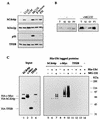
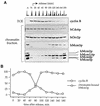
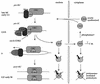
Evidence obtained from studies with yeast and Xenopus indicate that the initiation of DNA replication is a multistep process. The origin recognition complex (ORC), Cdc6p, and minichromosome maintenance (MCM) proteins are required for establishing prereplication complexes, upon which initiation is triggered by the activation of cyclin-dependent kinases and the Dbf4p-dependent kinase Cdc7p. The identification of human homologues of these replication proteins allows investigation of S-phase regulation in mammalian cells. Using centrifugal elutriation of several human cell lines, we demonstrate that whereas human Orc2 (hOrc2p) and hMcm proteins are present throughout the cell cycle, hCdc6p levels vary, being very low in early G(1) and accumulating until cells enter mitosis. hCdc6p can be polyubiquitinated in vivo, and it is stabilized by proteasome inhibitors. Similar to the case for hOrc2p, a significant fraction of hCdc6p is present on chromatin throughout the cell cycle, whereas hMcm proteins alternate between soluble and chromatin-bound forms. Loading of hMcm proteins onto chromatin occurs in late mitosis concomitant with the destruction of cyclin B, indicating that the mitotic kinase activity inhibits prereplication complex formation in human cells.
Figures



References
-
- Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
-
- Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. - PubMed
-
- Bolt M W, Mahoney P A. High-efficiency blotting of proteins of diverse sizes following sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Anal Biochem. 1997;247:185–192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
