The structure of the chromophore within DsRed, a red fluorescent protein from coral
- PMID: 11050230
- PMCID: PMC17282
- DOI: 10.1073/pnas.97.22.11990
The structure of the chromophore within DsRed, a red fluorescent protein from coral
Abstract
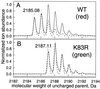
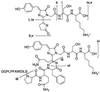
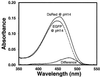
DsRed, a brilliantly red fluorescent protein, was recently cloned from Discosoma coral by homology to the green fluorescent protein (GFP) from the jellyfish Aequorea. A core question in the biochemistry of DsRed is the mechanism by which the GFP-like 475-nm excitation and 500-nm emission maxima of immature DsRed are red-shifted to the 558-nm excitation and 583-nm emission maxima of mature DsRed. After digestion of mature DsRed with lysyl endopeptidase, high-resolution mass spectra of the purified chromophore-bearing peptide reveal that some of the molecules have lost 2 Da relative to the peptide analogously prepared from a mutant, K83R, that stays green. Tandem mass spectrometry indicates that the bond between the alpha-carbon and nitrogen of Gln-66 has been dehydrogenated in DsRed, extending the GFP chromophore by forming C==N==C==O at the 2-position of the imidazolidinone. This acylimine substituent quantitatively accounts for the red shift according to quantum mechanical calculations. Reversible hydration of the C==N bond in the acylimine would explain why denaturation shifts mature DsRed back to a GFP-like absorbance. The C==N bond hydrolyses upon boiling, explaining why DsRed shows two fragment bands on SDS/PAGE. This assay suggests that conversion from green to red chromophores remains incomplete even after prolonged aging.
Figures

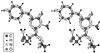

References
-
- Matz M V, Fradkov A F, Labas Y A, Savitsky A P, Zaraisky A G, Markelov M L, Lukyanov S A. Nat Biotechnol. 1999;17:969–973. - PubMed
-
- Tsien R Y. Nat Biotechnol. 1999;17:956–957. - PubMed
-
- Frisch M J, Trucks G W, Schlegel H B, Scuseria G E, Robb M A, Cheeseman J R, Zakrzewski V G, Montgomery J A, Jr, Stratmann R E, Burant J C, et al. gaussian 98 Revision A.7. Pittsburgh: Gaussian; 1998.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

