Replication by a single DNA polymerase of a stretched single-stranded DNA
- PMID: 11050232
- PMCID: PMC17284
- DOI: 10.1073/pnas.97.22.12002
Replication by a single DNA polymerase of a stretched single-stranded DNA
Abstract
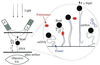
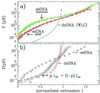
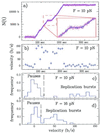
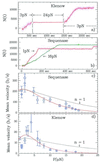
A new approach to the study of DNA/protein interactions has been opened through the recent advances in the manipulation of single DNA molecules. These allow the behavior of individual molecular motors to be studied under load and compared with bulk measurements. One example of such a motor is the DNA polymerase, which replicates DNA. We measured the replication rate by a single enzyme of a stretched single strand of DNA. The marked difference between the elasticity of single- and double-stranded DNA allows for the monitoring of replication in real time. We have found that the rate of replication depends strongly on the stretching force applied to the template. In particular, by varying the load we determined that the biochemical steps limiting replication are coupled to movement. The replication rate increases at low forces, decreases at forces greater than 4 pN, and ceases when the single-stranded DNA substrate is under a load greater than approximately 20 pN. The decay of the replication rate follows an Arrhenius law and indicates that multiple bases on the template strand are involved in the rate-limiting step of each cycle. This observation is consistent with the induced-fit mechanism for error detection during replication.
Figures

References
-
- Kornberg A, Baker T. DNA Replication. San Francisco: Freeman; 1992.
-
- Kunkel T A, Wilson S H. Nat Struct Biol. 1998;5:95–99. - PubMed
-
- Tabor S, Huber H E, Richardson C C. J Biol Chem. 1987;262:16212–16223. - PubMed
-
- Patel S S, Wong I, Johnson K A. Biochemistry. 1991;30:511–525. - PubMed
-
- Dahlberg M E, Bencovic S J. Biochemistry. 1991;30:4835–4843. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

