Maltose and maltodextrin transport in the thermoacidophilic gram-positive bacterium Alicyclobacillus acidocaldarius is mediated by a high-affinity transport system that includes a maltose binding protein tolerant to low pH
- PMID: 11053372
- PMCID: PMC94774
- DOI: 10.1128/JB.182.22.6292-6301.2000
Maltose and maltodextrin transport in the thermoacidophilic gram-positive bacterium Alicyclobacillus acidocaldarius is mediated by a high-affinity transport system that includes a maltose binding protein tolerant to low pH
Abstract
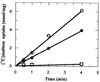
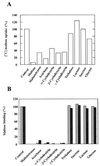
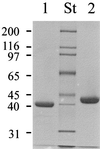
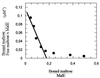
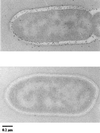
We have studied the uptake of maltose in the thermoacidophilic gram-positive bacterium Alicyclobacillus acidocaldarius, which grows best at 57 degrees C and pH 3.5. Under these conditions, accumulation of [(14)C]maltose was observed in cells grown with maltose but not in those grown with glucose. At lower temperatures or higher pH values, the transport rates substantially decreased. Uptake of radiolabeled maltose was inhibited by maltotetraose, acarbose, and cyclodextrins but not by lactose, sucrose, or trehalose. The kinetic parameters (K(m) of 0.91 +/- 0.06 microM and V(max) ranging from 0.6 to 3.7 nmol/min/mg of protein) are consistent with a binding protein-dependent ATP binding cassette (ABC) transporter. A corresponding binding protein (MalE) that interacts with maltose with high affinity (K(d) of 1.5 microM) was purified from the culture supernatant of maltose-grown cells. Immunoelectron microscopy revealed distribution of the protein throughout the cell wall. The malE gene was cloned and sequenced. Five additional open reading frames, encoding components of a maltose transport system (MalF and MalG), a putative transcriptional regulator (MalR), a cyclodextrinase (CdaA), and an alpha-glucosidase (GlcA), were identified downstream of malE. The malE gene lacking the DNA sequence that encodes the signal sequence was expressed in Escherichia coli. The purified wild-type and recombinant proteins bind maltose with high affinity over a wide pH range (2.5 to 7) and up to 80 degrees C. Recombinant MalE cross-reacted with an antiserum raised against the wild-type protein, thereby indicating that the latter is the product of the malE gene. The MalE protein might be well suited as a model to study tolerance of proteins to low pH.
Figures




References
-
- Bakker E P. The role of alkali-cation transport in energy coupling of neutrophilic and acidophilic bacteria: an assessment of methods and concepts. FEMS Microbiol Rev. 1990;75:319–334.
-
- Boos W, Lucht J M. Periplasmic binding-protein-dependent ABC-transporters. In: Neidthard F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1175–1209.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

