Bacillus subtilis SMC is required for proper arrangement of the chromosome and for efficient segregation of replication termini but not for bipolar movement of newly duplicated origin regions
- PMID: 11053392
- PMCID: PMC94794
- DOI: 10.1128/JB.182.22.6463-6471.2000
Bacillus subtilis SMC is required for proper arrangement of the chromosome and for efficient segregation of replication termini but not for bipolar movement of newly duplicated origin regions
Abstract
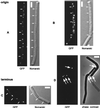
SMC protein is required for chromosome condensation and for the faithful segregation of daughter chromosomes in Bacillus subtilis. The visualization of specific sites on the chromosome showed that newly duplicated origin regions in growing cells of an smc mutant were able to segregate from each other but that the location of origin regions was frequently aberrant. In contrast, the segregation of replication termini was impaired in smc mutant cells. This analysis was extended to germinating spores of an smc mutant. The results showed that during germination, newly duplicated origins, but not termini, were able to separate from each other in the absence of SMC. Also, DAPI (4',6'-diamidino-2-phenylindole) staining revealed that chromosomes in germinating spores were able to undergo partial or complete replication but that the daughter chromosomes were blocked at a late stage in the segregation process. These findings were confirmed by time-lapse microscopy, which showed that after duplication in growing cells the origin regions underwent rapid movement toward opposite poles of the cell in the absence of SMC. This indicates that SMC is not a required component of the mitotic motor that initially drives origins apart after their duplication. It is also concluded that SMC is needed to maintain the proper layout of the chromosome in the cell and that it functions in the cell cycle after origin separation but prior to complete segregation or replication of daughter chromosomes. It is proposed here that chromosome segregation takes place in at least two steps: an SMC-independent step in which origins move apart and a subsequent SMC-dependent step in which newly duplicated chromosomes condense and are thereby drawn apart.
Figures





References
-
- Akhmedov A T, Frei C, Tsai-Pflugfelder M, Kemper B, Gasser S M, Jessberger R. Structural maintenance of chromosomes protein C-terminal domains bind preferentially to DNA with secondary structure. J Biol Chem. 1998;273:24088–24094. - PubMed
-
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. - PubMed
-
- Callister H, Wake R G. Completion of the replication and division cycle in temperature-sensitive DNA initiation mutants of Bacillus subtilis 168 at the non-permissive temperature. J Mol Biol. 1977;117:71–84. - PubMed
-
- Glaser P, Sharpe M E, Raether B, Perego M, Ohlsen K, Errington J. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 1997;11:1160–1168. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

