Escherichia coli TehB requires S-adenosylmethionine as a cofactor to mediate tellurite resistance
- PMID: 11053398
- PMCID: PMC94800
- DOI: 10.1128/JB.182.22.6509-6513.2000
Escherichia coli TehB requires S-adenosylmethionine as a cofactor to mediate tellurite resistance
Abstract
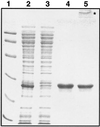
The Escherichia coli chromosomal determinant for tellurite resistance consists of two genes (tehA and tehB) which, when expressed on a multicopy plasmid, confer resistance to K(2)TeO(3) at 128 microg/ml, compared to the MIC of 2 microg/ml for the wild type. TehB is a cytoplasmic protein which possesses three conserved motifs (I, II, and III) found in S-adenosyl-L-methionine (SAM)-dependent non-nucleic acid methyltransferases. Replacement of the conserved aspartate residue in motif I by asparagine or alanine, or of the conserved phenylalanine in motif II by tyrosine or alanine, decreased resistance to background levels. Our results are consistent with motifs I and II in TehB being involved in SAM binding. Additionally, conformational changes in TehB are observed upon binding of both tellurite and SAM. The hydrodynamic radius of TehB measured by dynamic light scattering showed a approximately 20% decrease upon binding of both tellurite and SAM. These data suggest that TehB utilizes a methyltransferase activity in the detoxification of tellurite.
Figures
Similar articles
-
Crystallization and initial X-ray diffraction analysis of the tellurite-resistance S-adenosyl-L-methionine transferase protein TehB from Escherichia coli.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Nov 1;66(Pt 11):1496-9. doi: 10.1107/S1744309110036043. Epub 2010 Oct 28. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010. PMID: 21045305 Free PMC article.
-
The role of cysteine residues in tellurite resistance mediated by the TehAB determinant.Biochem Biophys Res Commun. 2000 Oct 22;277(2):394-400. doi: 10.1006/bbrc.2000.3686. Biochem Biophys Res Commun. 2000. PMID: 11032735
-
Characterization of gram-positive tellurite resistance encoded by the Streptococcus pneumoniae tehB gene.FEMS Microbiol Lett. 1999 May 15;174(2):385-92. doi: 10.1111/j.1574-6968.1999.tb13594.x. FEMS Microbiol Lett. 1999. PMID: 10339832
-
Increased resistance against tellurite is conferred by a mutation in the promoter region of uncommon tellurite resistance gene tehB in the ter-negative Shiga toxin-producing Escherichia coli O157:H7.Appl Environ Microbiol. 2024 Jun 18;90(6):e0228323. doi: 10.1128/aem.02283-23. Epub 2024 May 17. Appl Environ Microbiol. 2024. PMID: 38757978 Free PMC article.
-
Location of a potassium tellurite resistance operon (tehA tehB) within the terminus of Escherichia coli K-12.J Bacteriol. 1994 May;176(9):2740-2. doi: 10.1128/jb.176.9.2740-2742.1994. J Bacteriol. 1994. PMID: 8169225 Free PMC article.
Cited by
-
Metabolic Requirements of Escherichia coli in Intracellular Bacterial Communities during Urinary Tract Infection Pathogenesis.mBio. 2016 Apr 12;7(2):e00104-16. doi: 10.1128/mBio.00104-16. mBio. 2016. PMID: 27073089 Free PMC article.
-
In vivo 31P nuclear magnetic resonance investigation of tellurite toxicity in Escherichia coli.Appl Environ Microbiol. 2004 Dec;70(12):7342-7. doi: 10.1128/AEM.70.12.7342-7347.2004. Appl Environ Microbiol. 2004. PMID: 15574934 Free PMC article.
-
The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors.EMBO J. 2002 Feb 15;21(4):769-78. doi: 10.1093/emboj/21.4.769. EMBO J. 2002. PMID: 11847124 Free PMC article.
-
Crystallization and initial X-ray diffraction analysis of the tellurite-resistance S-adenosyl-L-methionine transferase protein TehB from Escherichia coli.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Nov 1;66(Pt 11):1496-9. doi: 10.1107/S1744309110036043. Epub 2010 Oct 28. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010. PMID: 21045305 Free PMC article.
-
DmsD, a Tat system specific chaperone, interacts with other general chaperones and proteins involved in the molybdenum cofactor biosynthesis.Biochim Biophys Acta. 2010 Jun;1804(6):1301-9. doi: 10.1016/j.bbapap.2010.01.022. Epub 2010 Feb 11. Biochim Biophys Acta. 2010. PMID: 20153451 Free PMC article.
References
-
- Amdur M L. Tellurium dioxide, an animal study in acute toxicity. AMA Arch Ind Health. 1958;17:665–667. - PubMed
-
- Bird M L, Challenger F. The formation of organo-metalloidal and similar compounds by micro-organisms. II. Dimethyltelluride. J Chem Soc. 1939;1939:163–168.
-
- Bradley D E. Detection of tellurite-resistance determinants in IncP plasmids. J Gen Microbiol. 1985;131:3135–3137. - PubMed
-
- Brady J M, Tobin J F, Gadd G M. Volatilization of selenite in an aqueous medium by a Penicillium species. Mycol Res. 1996;100:955–961.
-
- Carlton W W, Kelly W A. Tellurium toxicosis in Pekin ducks. Toxicol Appl Pharmacol. 1967;11:203–214.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

