Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related dinoflagellates
- PMID: 11055905
- PMCID: PMC92361
- DOI: 10.1128/AEM.66.11.4641-4648.2000
Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related dinoflagellates
Abstract
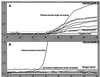
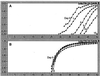
Pfiesteria complex species are heterotrophic and mixotrophic dinoflagellates that have been recognized as harmful algal bloom species associated with adverse fish and human health effects along the East Coast of North America, particularly in its largest (Chesapeake Bay in Maryland) and second largest (Albermarle-Pamlico Sound in North Carolina) estuaries. In response to impacts on human health and the economy, monitoring programs to detect the organism have been implemented in affected areas. However, until recently, specific identification of the two toxic species known thus far, Pfiesteria piscicida and P. shumwayae (sp. nov.), required scanning electron microscopy (SEM). SEM is a labor-intensive process in which a small number of cells can be analyzed, posing limitations when the method is applied to environmental estuarine water samples. To overcome these problems, we developed a real-time PCR-based assay that permits rapid and specific identification of these organisms in culture and heterogeneous environmental water samples. Various factors likely to be encountered when assessing environmental samples were addressed, and assay specificity was validated through screening of a comprehensive panel of cultures, including the two recognized Pfiesteria species, morphologically similar species, and a wide range of other estuarine dinoflagellates. Assay sensitivity and sample stability were established for both unpreserved and fixative (acidic Lugol's solution)-preserved samples. The effects of background DNA on organism detection and enumeration were also explored, and based on these results, we conclude that the assay may be utilized to derive quantitative data. This real-time PCR-based method will be useful for many other applications, including adaptation for field-based technology.
Figures




References
-
- Burkholder J M, Hobbs C W, Glasgow H B., Jr Distribution and environmental conditions for fish kills linked to a toxic ambush-predator dinoflagellate. Mar Ecol Prog Ser. 1995;124:43–61.
-
- Burkholder J M. Implications of harmful marine microalgae and heterotrophic dinoflagellates in management of sustainable marine fisheries. Ecol Applications. 1998;8(Suppl.):S37–S62.
-
- Burkholder, J. M. Critical needs in harmful algal bloom research, p. 232–269. In S. Vaupel (ed.), Proceedings of the Workshop on Opportunities for Environmental Application of Marine Biotechnology, in press. National Academy of Sciences, Washington, D.C.
-
- Burkholder J M, Glasgow H B., Jr Interactions of a toxic estuarine dinoflagellate with microbial predators and prey. Arch Protistenkd. 1995;145:177–188.
-
- Burkholder J M, Glasgow H B., Jr Pfiesteria piscicida and other toxic Pfiesteria-like dinoflagellates: behavior, impacts, and environmental controls. Limnol Oceanogr. 1997;42:1052–1075.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

