Key aromatic-ring-cleaving enzyme, protocatechuate 3,4-dioxygenase, in the ecologically important marine Roseobacter lineage
- PMID: 11055908
- PMCID: PMC92364
- DOI: 10.1128/AEM.66.11.4662-4672.2000
Key aromatic-ring-cleaving enzyme, protocatechuate 3,4-dioxygenase, in the ecologically important marine Roseobacter lineage
Abstract
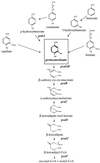
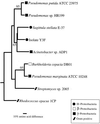
Aromatic compound degradation in six bacteria representing an ecologically important marine taxon of the alpha-proteobacteria was investigated. Initial screens suggested that isolates in the Roseobacter lineage can degrade aromatic compounds via the beta-ketoadipate pathway, a catabolic route that has been well characterized in soil microbes. Six Roseobacter isolates were screened for the presence of protocatechuate 3,4-dioxygenase, a key enzyme in the beta-ketoadipate pathway. All six isolates were capable of growth on at least three of the eight aromatic monomers presented (anthranilate, benzoate, p-hydroxybenzoate, salicylate, vanillate, ferulate, protocatechuate, and coumarate). Four of the Roseobacter group isolates had inducible protocatechuate 3, 4-dioxygenase activity in cell extracts when grown on p-hydroxybenzoate. The pcaGH genes encoding this ring cleavage enzyme were cloned and sequenced from two isolates, Sagittula stellata E-37 and isolate Y3F, and in both cases the genes could be expressed in Escherichia coli to yield dioxygenase activity. Additional genes involved in the protocatechuate branch of the beta-ketoadipate pathway (pcaC, pcaQ, and pobA) were found to cluster with pcaGH in these two isolates. Pairwise sequence analysis of the pca genes revealed greater similarity between the two Roseobacter group isolates than between genes from either Roseobacter strain and soil bacteria. A degenerate PCR primer set targeting a conserved region within PcaH successfully amplified a fragment of pcaH from two additional Roseobacter group isolates, and Southern hybridization indicated the presence of pcaH in the remaining two isolates. This evidence of protocatechuate 3, 4-dioxygenase and the beta-ketoadipate pathway was found in all six Roseobacter isolates, suggesting widespread abilities to degrade aromatic compounds in this marine lineage.
Figures



References
-
- Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Dagley S. Biochemistry of aromatic hydrocarbon degradation in pseudomonads. In: Sokatch J R, editor. The bacteria. Vol. 10. New York, N.Y: Academic Press, Inc.; 1986. pp. 527–555.
-
- Delneri D, Degrassi G, Rizzo R, Bruschi C V. Degradation of trans-ferulic acid and p-courmaric acid by Acinetobacter calcoaceticus DSM 586. Biochim Biophys Acta. 1995;1244:363–367. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

