Identification and characterization of an ATP binding cassette L-carnitine transporter in Listeria monocytogenes
- PMID: 11055912
- PMCID: PMC92368
- DOI: 10.1128/AEM.66.11.4696-4704.2000
Identification and characterization of an ATP binding cassette L-carnitine transporter in Listeria monocytogenes
Abstract
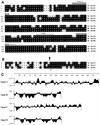
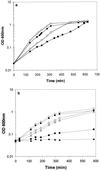
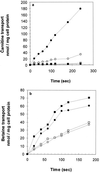
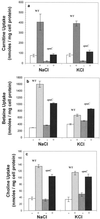
We identified an operon in Listeria monocytogenes EGD with high levels of sequence similarity to the operons encoding the OpuC and OpuB compatible solute transporters from Bacillus subtilis, which are members of the ATP binding cassette (ABC) substrate binding protein-dependent transporter superfamily. The operon, designated opuC, consists of four genes which are predicted to encode an ATP binding protein (OpuCA), an extracellular substrate binding protein (OpuCC), and two membrane-associated proteins presumed to form the permease (OpuCB and OpuCD). The operon is preceded by a potential SigB-dependent promoter. An opuC-defective mutant was generated by the insertional inactivation of the opuCA gene. The mutant was impaired for growth at high osmolarity in brain heart infusion broth and failed to grow in a defined medium. Supplementation of the defined medium with peptone restored the growth of the mutant in this medium. The mutant was found to accumulate the compatible solutes glycine betaine and choline to same extent as the parent strain but was defective in the uptake of L-carnitine. We conclude that the opuC operon in L. monocytogenes encodes an ABC compatible solute transporter which is capable of transporting L-carnitine and which plays an important role in osmoregulation in this pathogen.
Figures


Similar articles
-
Role of sigmaB in regulating the compatible solute uptake systems of Listeria monocytogenes: osmotic induction of opuC is sigmaB dependent.Appl Environ Microbiol. 2003 Apr;69(4):2015-22. doi: 10.1128/AEM.69.4.2015-2022.2003. Appl Environ Microbiol. 2003. PMID: 12676677 Free PMC article.
-
Molecular and physiological analysis of the role of osmolyte transporters BetL, Gbu, and OpuC in growth of Listeria monocytogenes at low temperatures.Appl Environ Microbiol. 2004 May;70(5):2912-8. doi: 10.1128/AEM.70.5.2912-2918.2004. Appl Environ Microbiol. 2004. PMID: 15128551 Free PMC article.
-
Identification of opuC as a chill-activated and osmotically activated carnitine transporter in Listeria monocytogenes.Appl Environ Microbiol. 2002 Jun;68(6):2644-50. doi: 10.1128/AEM.68.6.2644-2650.2002. Appl Environ Microbiol. 2002. PMID: 12039715 Free PMC article.
-
Identification and analysis of the osmotolerance associated genes in Listeria monocytogenes.Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2008 Sep;25(9):1089-94. doi: 10.1080/02652030802056634. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2008. PMID: 19238619 Review.
-
Biochemical and structural analysis of the Bacillus subtilis ABC transporter OpuA and its isolated subunits.J Mol Microbiol Biotechnol. 2005;10(2-4):76-91. doi: 10.1159/000091556. J Mol Microbiol Biotechnol. 2005. PMID: 16645306 Review.
Cited by
-
A single point mutation in the listerial betL σ(A)-dependent promoter leads to improved osmo- and chill-tolerance and a morphological shift at elevated osmolarity.Bioengineered. 2013 Nov-Dec;4(6):401-7. doi: 10.4161/bioe.24094. Epub 2013 Mar 11. Bioengineered. 2013. PMID: 23478432 Free PMC article.
-
The Flagellar Transcriptional Regulator FtcR Controls Brucella melitensis 16M Biofilm Formation via a betI-Mediated Pathway in Response to Hyperosmotic Stress.Int J Mol Sci. 2022 Aug 31;23(17):9905. doi: 10.3390/ijms23179905. Int J Mol Sci. 2022. PMID: 36077302 Free PMC article.
-
Characterization of the osmoprotectant transporter OpuC from Pseudomonas syringae and demonstration that cystathionine-beta-synthase domains are required for its osmoregulatory function.J Bacteriol. 2007 Oct;189(19):6901-12. doi: 10.1128/JB.00763-07. Epub 2007 Jul 27. J Bacteriol. 2007. PMID: 17660277 Free PMC article.
-
Antibacterial Efficacy and Mechanism of Mannosylerythritol Lipids-A on Listeria monocytogenes.Molecules. 2020 Oct 21;25(20):4857. doi: 10.3390/molecules25204857. Molecules. 2020. PMID: 33096808 Free PMC article.
-
A Listeria monocytogenes RNA helicase essential for growth and ribosomal maturation at low temperatures uses its C terminus for appropriate interaction with the ribosome.J Bacteriol. 2012 Aug;194(16):4377-85. doi: 10.1128/JB.00348-12. Epub 2012 Jun 15. J Bacteriol. 2012. PMID: 22707705 Free PMC article.
References
-
- Amezaga M-R, Davidson I, McLaggan D, Verheul A, Abee T, Booth I R. The role of peptide metabolism in the growth of Listeria monocytogenes ATCC 23074 at high osmolarity. Microbiology. 1995;141:41–49. - PubMed
-
- Boos W, Lucht J M. Periplasmic binding protein dependent ABC transporters. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1175–1209.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

