Complete nucleotide sequence of ubiquitous plasmid pEA29 from Erwinia amylovora strain Ea88: gene organization and intraspecies variation
- PMID: 11055941
- PMCID: PMC92397
- DOI: 10.1128/AEM.66.11.4897-4907.2000
Complete nucleotide sequence of ubiquitous plasmid pEA29 from Erwinia amylovora strain Ea88: gene organization and intraspecies variation
Abstract
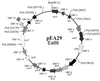
The complete sequence of plasmid pEA29 from Erwinia amylovora strain Ea88 consists of 28,185 bp with a 50.2% G+C content. As deletions and insertions were detected in other derivatives of pEA29, its size actually varied from 27.6 to 34.9 kb. Thirteen open reading frames that encoded predicted proteins with similarities to known proteins from other bacteria were identified along with two open reading frames related to hypothetical proteins found in GenBank and six open reading frames with no similarities to existing GenBank entries. Predicted products of open reading frames with similarity to the thiamine biosynthetic genes thiO, thiG, and thiF; a betT gene coding for choline transport; an msrA gene for the enzyme methionine sulfoxide reductase; a putative methyl-accepting chemotaxis gene; an aldehyde dehydrogenase gene; an hns DNA binding gene; a LysR-type transcriptional regulator; and parA and parB partitioning genes were identified. A putative iteron-containing theta-type origin of replication with an AT-rich region and a gene for a RepA protein was identified. PstI and KpnI restriction patterns for pEA29 isolated from tree fruit strains of E. amylovora were homogenous and different from those for pEA29 isolated from Rubus (raspberry) strains. All Rubus derivatives of pEA29 contained a point mutation that eliminated a PstI site and a 1,264-bp region that replaced 1, 890 bp of sequence found in pEA29 from strain Ea88. This change eliminated a second PstI site and increased the length of a KpnI fragment. An insertion sequence, ISEam1, was detected in one Rubus strain, and transposon Tn5393 was detected in three apple strains in two separate locations on the plasmid. Plasmid-cured strains exhibited reduced virulence and modified colony morphology on minimal medium without thiamine, indicating that some of the genes in pEA29 play a role in the physiology or metabolism of E. amylovora.
Figures


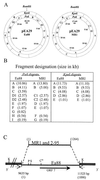
 ) indicate sequenced
regions of Tn2501. (A) Physical and genetic map of
Tn2501 compared to those of the partial res site
and resolvase (tnpR) gene found on pEA29 (nt 18084 to
18785). Positions for the transposase gene (tnpA),
res site, and resolvase gene (tnpR) of
Tn2501 are indicated by the hatched box, white box, and
latticework box, respectively. Gene orientation is indicated by arrows.
Restriction sites are as follows: BamHI (B),
BglII (Bg), HindIII (H), SalI (S),
SmaI (Sm), and SstI (Ss). Inverted repeats for
Tn2501 are indicated by IR-L (left) and IR-R (right). (B)
Sequence alignment of the res regions of Tn2501,
Tn2502, and pEA29 (nt 18084 to 18188). Dyad symmetry regions
consistent with Tn2501 and Tn2502 res sites I,
II, and III are boxed, as is the corresponding pEA29 sequence. Areas
where the sequence is in agreement with Tn2501 are indicated
by asterisks.
) indicate sequenced
regions of Tn2501. (A) Physical and genetic map of
Tn2501 compared to those of the partial res site
and resolvase (tnpR) gene found on pEA29 (nt 18084 to
18785). Positions for the transposase gene (tnpA),
res site, and resolvase gene (tnpR) of
Tn2501 are indicated by the hatched box, white box, and
latticework box, respectively. Gene orientation is indicated by arrows.
Restriction sites are as follows: BamHI (B),
BglII (Bg), HindIII (H), SalI (S),
SmaI (Sm), and SstI (Ss). Inverted repeats for
Tn2501 are indicated by IR-L (left) and IR-R (right). (B)
Sequence alignment of the res regions of Tn2501,
Tn2502, and pEA29 (nt 18084 to 18188). Dyad symmetry regions
consistent with Tn2501 and Tn2502 res sites I,
II, and III are boxed, as is the corresponding pEA29 sequence. Areas
where the sequence is in agreement with Tn2501 are indicated
by asterisks.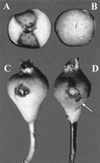

References
-
- Andresen P A, Kaasen I, Styrvold O B, Boulnois G, Strom A R. Molecular cloning, physical mapping and expression of the bet genes governing the osmoregulatory choline glycine betaine pathway of Escherichia coli. J Gen Microbiol. 1988;134:1737–1746. - PubMed
-
- Begley T P, Downs D M, Ealick S E, McLafferty F W, Van Loon A P G M, Taylor S, Campobasso N, Chiu H-J, Kinsland C, Reddick J J, Xi J. Thaimin biosynthesis in prokaryotes. Arch Microbiol. 1999;171:293–300. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

